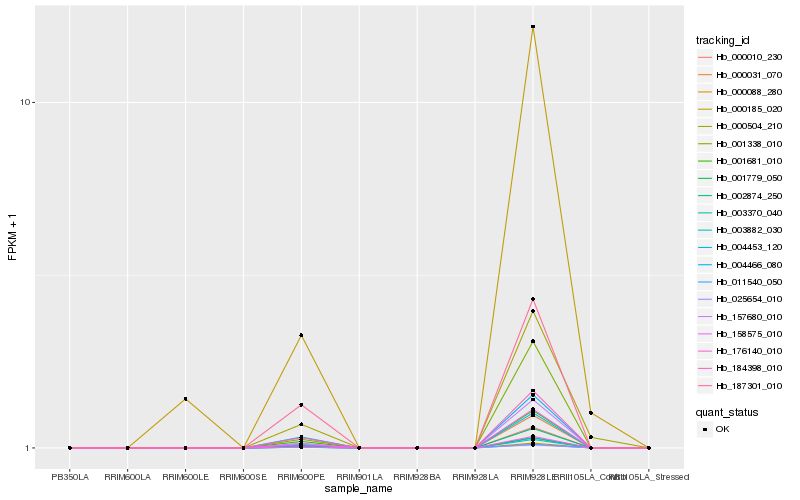
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000504_210 |
0.0 |
- |
- |
hypothetical protein JCGZ_10251 [Jatropha curcas] |
| 2 |
Hb_000185_020 |
0.1367737499 |
- |
- |
- |
| 3 |
Hb_000010_230 |
0.1467247193 |
- |
- |
PREDICTED: F-box/WD repeat-containing protein 11-like [Jatropha curcas] |
| 4 |
Hb_002874_250 |
0.1468623466 |
- |
- |
- |
| 5 |
Hb_003882_030 |
0.1473383319 |
- |
- |
putative retrotransposon protein [Phyllostachys edulis] |
| 6 |
Hb_004453_120 |
0.1504750385 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 7 |
Hb_157680_010 |
0.1511609611 |
- |
- |
PREDICTED: uncharacterized protein LOC103444007 [Malus domestica] |
| 8 |
Hb_158575_010 |
0.151310409 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_184398_010 |
0.1513460405 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 10 |
Hb_001681_010 |
0.1523419471 |
- |
- |
PREDICTED: cytochrome P450 81E8-like isoform X1 [Populus euphratica] |
| 11 |
Hb_004466_080 |
0.1585828326 |
- |
- |
PREDICTED: SNF1-related protein kinase regulatory subunit gamma-1-like [Jatropha curcas] |
| 12 |
Hb_176140_010 |
0.1591625551 |
- |
- |
polyprotein [Oryza australiensis] |
| 13 |
Hb_001779_050 |
0.1595218689 |
- |
- |
polyprotein [Ananas comosus] |
| 14 |
Hb_025654_010 |
0.1643477619 |
- |
- |
PREDICTED: uncharacterized protein LOC104221396 [Nicotiana sylvestris] |
| 15 |
Hb_001338_010 |
0.1648075742 |
transcription factor |
TF Family: GNAT |
GCN5-related N-acetyltransferase family protein [Populus trichocarpa] |
| 16 |
Hb_011540_050 |
0.1650323014 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 17 |
Hb_187301_010 |
0.165166765 |
- |
- |
PREDICTED: uncharacterized protein LOC105629082 [Jatropha curcas] |
| 18 |
Hb_000088_280 |
0.1667773411 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At3g47570 [Jatropha curcas] |
| 19 |
Hb_000031_070 |
0.1670022418 |
- |
- |
Thioredoxin domain-containing protein, putative [Ricinus communis] |
| 20 |
Hb_003370_040 |
0.1688805312 |
- |
- |
PREDICTED: uncharacterized protein LOC105629587 [Jatropha curcas] |