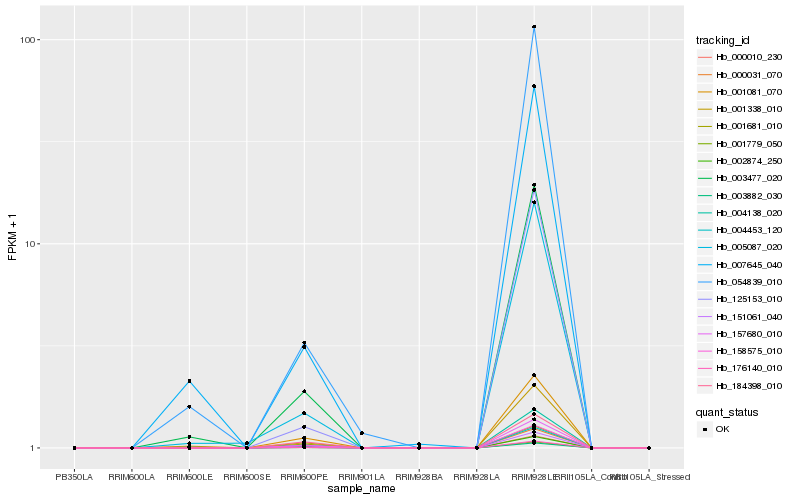
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_176140_010 |
0.0 |
- |
- |
polyprotein [Oryza australiensis] |
| 2 |
Hb_001779_050 |
0.0009244222 |
- |
- |
polyprotein [Ananas comosus] |
| 3 |
Hb_001338_010 |
0.0134773425 |
transcription factor |
TF Family: GNAT |
GCN5-related N-acetyltransferase family protein [Populus trichocarpa] |
| 4 |
Hb_000031_070 |
0.0182440468 |
- |
- |
Thioredoxin domain-containing protein, putative [Ricinus communis] |
| 5 |
Hb_158575_010 |
0.0244675223 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_157680_010 |
0.0250660414 |
- |
- |
PREDICTED: uncharacterized protein LOC103444007 [Malus domestica] |
| 7 |
Hb_004453_120 |
0.0279330234 |
- |
- |
ammonium transporter, putative [Ricinus communis] |
| 8 |
Hb_003882_030 |
0.0459694301 |
- |
- |
putative retrotransposon protein [Phyllostachys edulis] |
| 9 |
Hb_002874_250 |
0.0507247862 |
- |
- |
- |
| 10 |
Hb_000010_230 |
0.052479048 |
- |
- |
PREDICTED: F-box/WD repeat-containing protein 11-like [Jatropha curcas] |
| 11 |
Hb_003477_020 |
0.0654022295 |
desease resistance |
Gene Name: AAA_29 |
PREDICTED: ABC transporter G family member 15-like [Populus euphratica] |
| 12 |
Hb_151061_040 |
0.0712282439 |
- |
- |
putative retrotransposon protein [Phyllostachys edulis] |
| 13 |
Hb_004138_020 |
0.0844683263 |
- |
- |
PREDICTED: cucumisin-like [Populus euphratica] |
| 14 |
Hb_005087_020 |
0.0855148523 |
- |
- |
Metalloendoproteinase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_001081_070 |
0.0931232324 |
- |
- |
hypothetical protein POPTR_0001s08730g [Populus trichocarpa] |
| 16 |
Hb_184398_010 |
0.101893927 |
- |
- |
PREDICTED: putative ribonuclease H protein At1g65750-like [Citrus sinensis] |
| 17 |
Hb_001681_010 |
0.1056204689 |
- |
- |
PREDICTED: cytochrome P450 81E8-like isoform X1 [Populus euphratica] |
| 18 |
Hb_125153_010 |
0.1060975225 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD1-1 [Malus domestica] |
| 19 |
Hb_054839_010 |
0.1093163486 |
- |
- |
PsbC, partial (chloroplast) [Asimina parviflora] |
| 20 |
Hb_007645_040 |
0.1110740196 |
- |
- |
ribulose-1,5-bisphosphate carboxylase/oxygenase large subunit [Tachigali myrmecophila] |