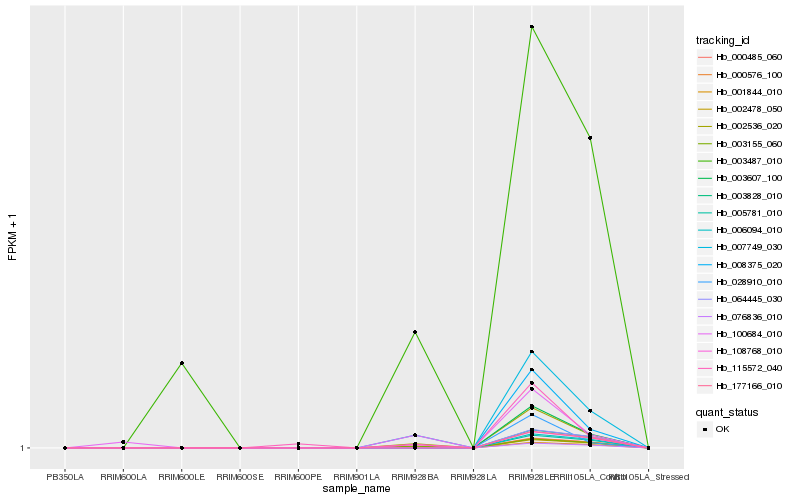
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002536_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105801112 [Gossypium raimondii] |
| 2 |
Hb_007749_030 |
0.0227802578 |
- |
- |
- |
| 3 |
Hb_003155_060 |
0.1330179352 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 4 |
Hb_000576_100 |
0.1333616595 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 5 |
Hb_108768_010 |
0.1377277886 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 6 |
Hb_003607_100 |
0.1890767282 |
- |
- |
PREDICTED: zinc finger protein 3 [Jatropha curcas] |
| 7 |
Hb_115572_040 |
0.1936746866 |
- |
- |
PREDICTED: uncharacterized protein LOC105632710 [Jatropha curcas] |
| 8 |
Hb_008375_020 |
0.1996086458 |
- |
- |
PREDICTED: uncharacterized protein LOC104894372 [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_003487_010 |
0.2016407802 |
- |
- |
- |
| 10 |
Hb_100684_010 |
0.2108274177 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 11 |
Hb_028910_010 |
0.2216302943 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 12 |
Hb_076836_010 |
0.2255088367 |
- |
- |
PREDICTED: uncharacterized protein LOC104801925 [Tarenaya hassleriana] |
| 13 |
Hb_001844_010 |
0.2257207462 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 14 |
Hb_002478_050 |
0.2265455307 |
- |
- |
K(+)/H(+) antiporter, putative [Ricinus communis] |
| 15 |
Hb_000485_060 |
0.227035285 |
- |
- |
PREDICTED: uncharacterized protein LOC105641351 [Jatropha curcas] |
| 16 |
Hb_005781_010 |
0.2279428055 |
- |
- |
PREDICTED: probable terpene synthase 6 [Jatropha curcas] |
| 17 |
Hb_006094_010 |
0.2283696094 |
- |
- |
PREDICTED: uncharacterized protein LOC104227769, partial [Nicotiana sylvestris] |
| 18 |
Hb_177166_010 |
0.2292076794 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 19 |
Hb_003828_010 |
0.229708621 |
- |
- |
PREDICTED: uncharacterized protein LOC104219776 [Nicotiana sylvestris] |
| 20 |
Hb_064445_030 |
0.2298175322 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |