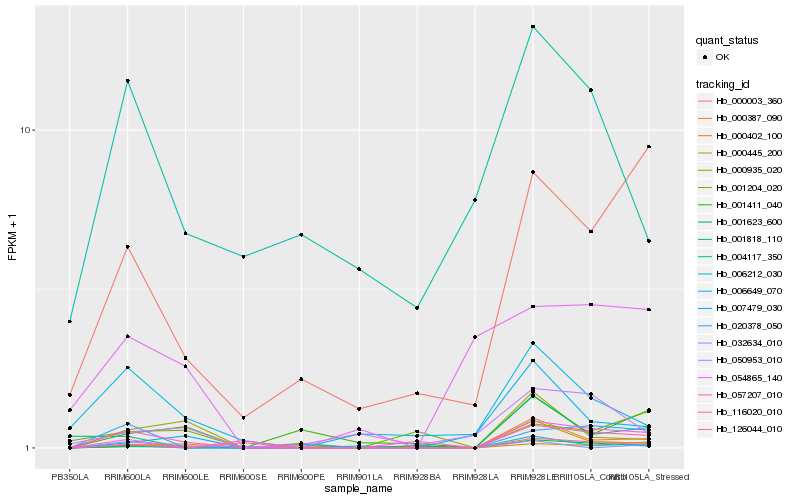
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_116020_010 |
0.0 |
- |
- |
Uncharacterized protein TCM_023639 [Theobroma cacao] |
| 2 |
Hb_020378_050 |
0.2298191793 |
- |
- |
- |
| 3 |
Hb_000387_090 |
0.2662324894 |
- |
- |
PREDICTED: uncharacterized protein LOC104115978 [Nicotiana tomentosiformis] |
| 4 |
Hb_000003_360 |
0.297181568 |
- |
- |
Putative inactive purple acid phosphatase 27 [Glycine soja] |
| 5 |
Hb_000935_020 |
0.3231446883 |
- |
- |
putative non-LTR retroelement reverse transcriptase [Arabidopsis thaliana] |
| 6 |
Hb_006649_070 |
0.3276340932 |
- |
- |
unknown [Lotus japonicus] |
| 7 |
Hb_001623_600 |
0.3303731127 |
- |
- |
PREDICTED: uncharacterized protein LOC104748784 [Camelina sativa] |
| 8 |
Hb_054865_140 |
0.3364781221 |
- |
- |
- |
| 9 |
Hb_000402_100 |
0.3537846289 |
- |
- |
PREDICTED: L-tryptophan--pyruvate aminotransferase 1 [Jatropha curcas] |
| 10 |
Hb_001818_110 |
0.367449192 |
- |
- |
- |
| 11 |
Hb_000445_200 |
0.3693463782 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 12 |
Hb_001204_020 |
0.3697804008 |
- |
- |
Uncharacterized protein TCM_021965 [Theobroma cacao] |
| 13 |
Hb_057207_010 |
0.3713044133 |
- |
- |
hypothetical protein F383_17010 [Gossypium arboreum] |
| 14 |
Hb_050953_010 |
0.3718446021 |
- |
- |
PREDICTED: uncharacterized protein LOC105779031 [Gossypium raimondii] |
| 15 |
Hb_001411_040 |
0.3726642247 |
- |
- |
hypothetical protein VITISV_010644 [Vitis vinifera] |
| 16 |
Hb_126044_010 |
0.3782131779 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 17 |
Hb_006212_030 |
0.3836571777 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 18 |
Hb_007479_030 |
0.3879753872 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 3 [Jatropha curcas] |
| 19 |
Hb_032634_010 |
0.3888592001 |
- |
- |
PREDICTED: uncharacterized protein LOC104216022, partial [Nicotiana sylvestris] |
| 20 |
Hb_004117_350 |
0.3920065929 |
transcription factor |
TF Family: OFP |
PREDICTED: transcription repressor OFP4 [Jatropha curcas] |