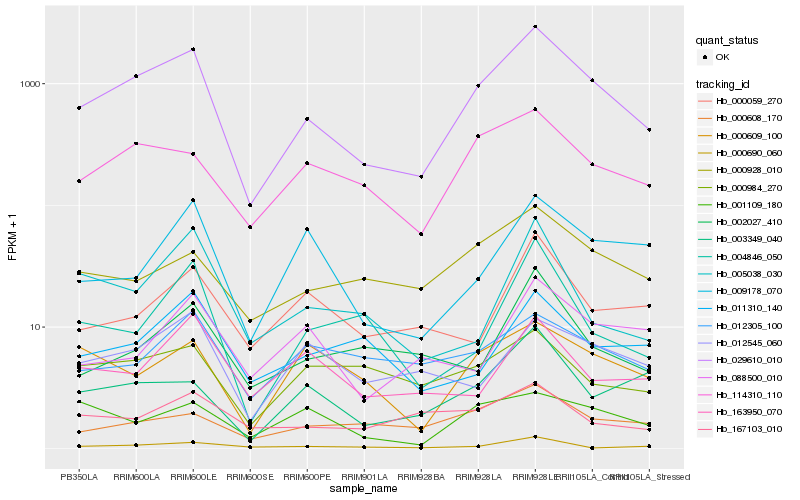
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_029610_010 |
0.0 |
- |
- |
caffeic acid 3-O-methyltransferase [Hevea brasiliensis] |
| 2 |
Hb_114310_110 |
0.169305153 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 3 |
Hb_000928_010 |
0.1900274678 |
- |
- |
Cell division protein ftsZ, putative [Ricinus communis] |
| 4 |
Hb_000608_170 |
0.1925032508 |
- |
- |
F23N19.4 [Arabidopsis thaliana] |
| 5 |
Hb_003349_040 |
0.1965887283 |
- |
- |
PREDICTED: uncharacterized protein LOC105628285 [Jatropha curcas] |
| 6 |
Hb_009178_070 |
0.1972243569 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 8, chloroplastic [Jatropha curcas] |
| 7 |
Hb_012545_060 |
0.2027009684 |
- |
- |
PREDICTED: uncharacterized protein LOC103986927 [Musa acuminata subsp. malaccensis] |
| 8 |
Hb_000609_100 |
0.2030145214 |
- |
- |
Thylakoid lumenal 16.5 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_000690_060 |
0.2031740333 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Populus euphratica] |
| 10 |
Hb_011310_140 |
0.2058093581 |
- |
- |
hypothetical protein JCGZ_07173 [Jatropha curcas] |
| 11 |
Hb_002027_410 |
0.2062611423 |
- |
- |
PREDICTED: protease Do-like 2, chloroplastic [Jatropha curcas] |
| 12 |
Hb_004846_050 |
0.2065253614 |
- |
- |
PREDICTED: uncharacterized protein ycf23-like [Jatropha curcas] |
| 13 |
Hb_001109_180 |
0.2080289605 |
- |
- |
PREDICTED: protein BREVIS RADIX-like [Populus euphratica] |
| 14 |
Hb_012305_100 |
0.2081910328 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: dihydrolipoyl dehydrogenase 2, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_000984_270 |
0.2084387616 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000059_270 |
0.2085143457 |
- |
- |
superoxide dismutase [Fe], chloroplastic [Jatropha curcas] |
| 17 |
Hb_167103_010 |
0.2106798838 |
- |
- |
PREDICTED: transcriptional activator DEMETER isoform X1 [Jatropha curcas] |
| 18 |
Hb_005038_030 |
0.2115579413 |
- |
- |
PREDICTED: uncharacterized protein LOC105636682 isoform X2 [Jatropha curcas] |
| 19 |
Hb_163950_070 |
0.2115715104 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 20 |
Hb_088500_010 |
0.2125046312 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At4g39970 [Jatropha curcas] |