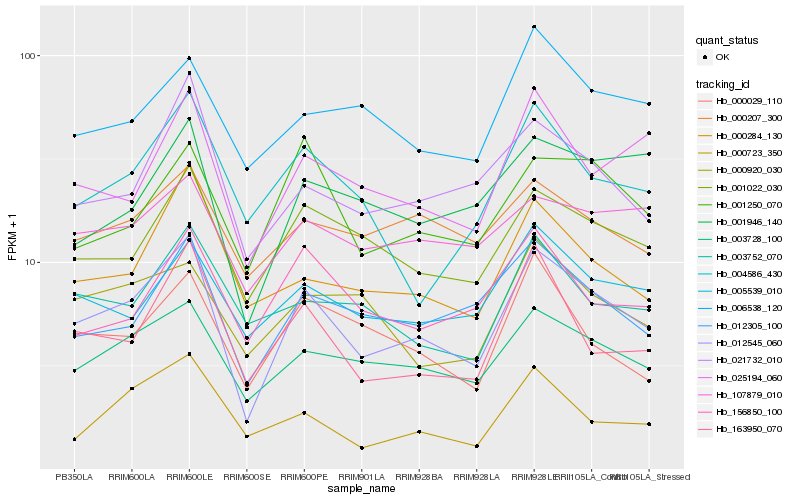
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_012545_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC103986927 [Musa acuminata subsp. malaccensis] |
| 2 |
Hb_003728_100 |
0.1118812528 |
transcription factor |
TF Family: FAR1 |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001022_030 |
0.1239613821 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 4 |
Hb_012305_100 |
0.1258509553 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
PREDICTED: dihydrolipoyl dehydrogenase 2, chloroplastic-like [Jatropha curcas] |
| 5 |
Hb_025194_060 |
0.1301568735 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 5, chloroplastic isoform X2 [Jatropha curcas] |
| 6 |
Hb_021732_010 |
0.1314636826 |
- |
- |
biotin carboxylase [Vernicia fordii] |
| 7 |
Hb_163950_070 |
0.1319985014 |
- |
- |
arginine/serine-rich splicing factor, putative [Ricinus communis] |
| 8 |
Hb_000284_130 |
0.1336614558 |
- |
- |
sodium-bile acid cotransporter, putative [Ricinus communis] |
| 9 |
Hb_005539_010 |
0.1389440289 |
- |
- |
PREDICTED: uncharacterized protein LOC105644585 [Jatropha curcas] |
| 10 |
Hb_000029_110 |
0.1409787545 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001250_070 |
0.1434883688 |
- |
- |
ATP-citrate synthase, putative [Ricinus communis] |
| 12 |
Hb_001946_140 |
0.1441387046 |
- |
- |
PREDICTED: heme oxygenase 1, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_004586_430 |
0.1441473271 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000723_350 |
0.1459412813 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 15 |
Hb_156850_100 |
0.1475332172 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 16 |
Hb_003752_070 |
0.1483031765 |
- |
- |
PREDICTED: uncharacterized protein LOC105643912 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000207_300 |
0.149856916 |
rubber biosynthesis |
Gene Name: Dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex |
PREDICTED: dihydrolipoyllysine-residue acetyltransferase component 5 of pyruvate dehydrogenase complex, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_006538_120 |
0.1501274943 |
- |
- |
PREDICTED: 50S ribosomal protein L15, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000920_030 |
0.151592812 |
- |
- |
short chain alcohol dehydrogenase, putative [Ricinus communis] |
| 20 |
Hb_107879_010 |
0.1519311871 |
- |
- |
phosphoglycerate mutase, putative [Ricinus communis] |