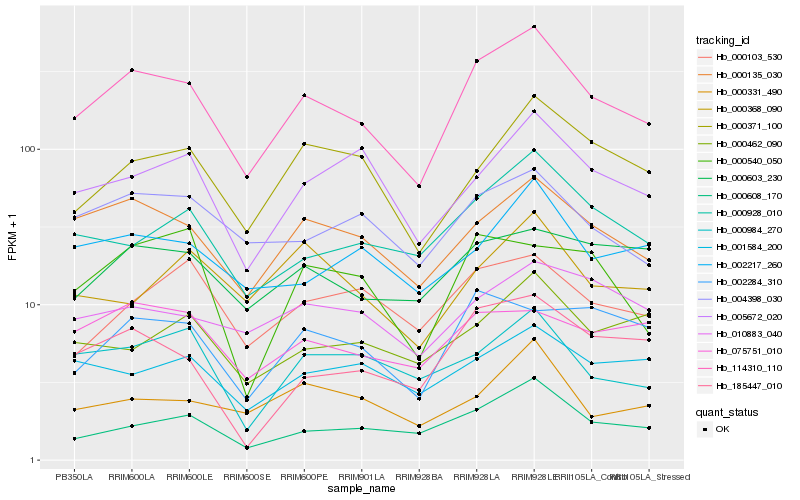
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_114310_110 |
0.0 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 2 |
Hb_000135_030 |
0.1114029545 |
- |
- |
PREDICTED: uncharacterized protein LOC105634729 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000608_170 |
0.1223363859 |
- |
- |
F23N19.4 [Arabidopsis thaliana] |
| 4 |
Hb_004398_030 |
0.1358018203 |
- |
- |
PREDICTED: chloroplast processing peptidase [Jatropha curcas] |
| 5 |
Hb_000928_010 |
0.137863665 |
- |
- |
Cell division protein ftsZ, putative [Ricinus communis] |
| 6 |
Hb_000371_100 |
0.1391071815 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
RecName: Full=Geranylgeranyl pyrophosphate synthase, chloroplastic; Short=GGPP synthase; Short=GGPS; AltName: Full=(2E,6E)-farnesyl diphosphate synthase; AltName: Full=Dimethylallyltranstransferase; AltName: Full=Farnesyl diphosphate synthase; AltName: Full=Farnesyltranstransferase; AltName: Full=Geranyltranstransferase; Flags: Precursor [Hevea brasiliensis] |
| 7 |
Hb_185447_010 |
0.1445431219 |
- |
- |
hypothetical protein JCGZ_13512 [Jatropha curcas] |
| 8 |
Hb_005672_020 |
0.1460878267 |
- |
- |
PREDICTED: 50S ribosomal protein L6, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002217_260 |
0.1467713137 |
- |
- |
PREDICTED: uncharacterized protein LOC105643577 [Jatropha curcas] |
| 10 |
Hb_000603_230 |
0.1493846425 |
- |
- |
copper-transporting atpase paa1, putative [Ricinus communis] |
| 11 |
Hb_000984_270 |
0.1499575435 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000331_490 |
0.1530439648 |
- |
- |
PREDICTED: radical S-adenosyl methionine domain-containing protein 1, mitochondrial [Jatropha curcas] |
| 13 |
Hb_010883_040 |
0.1534806553 |
- |
- |
PREDICTED: phosphatidylinositol 4-phosphate 5-kinase 1 [Jatropha curcas] |
| 14 |
Hb_002284_310 |
0.1549090018 |
- |
- |
PREDICTED: uncharacterized protein LOC105635220 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001584_200 |
0.157275777 |
- |
- |
PREDICTED: Golgi apparatus membrane protein-like protein ECHIDNA [Jatropha curcas] |
| 16 |
Hb_075751_010 |
0.1577594816 |
- |
- |
PREDICTED: uncharacterized protein LOC105635783 isoform X2 [Jatropha curcas] |
| 17 |
Hb_000103_530 |
0.1583810548 |
- |
- |
PREDICTED: uncharacterized protein LOC105632061 [Jatropha curcas] |
| 18 |
Hb_000462_090 |
0.1587892085 |
- |
- |
PREDICTED: carotene epsilon-monooxygenase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000368_090 |
0.1603812526 |
- |
- |
TPA: hypothetical protein ZEAMMB73_442906 [Zea mays] |
| 20 |
Hb_000540_050 |
0.1653571222 |
- |
- |
PREDICTED: 125 kDa kinesin-related protein-like [Jatropha curcas] |