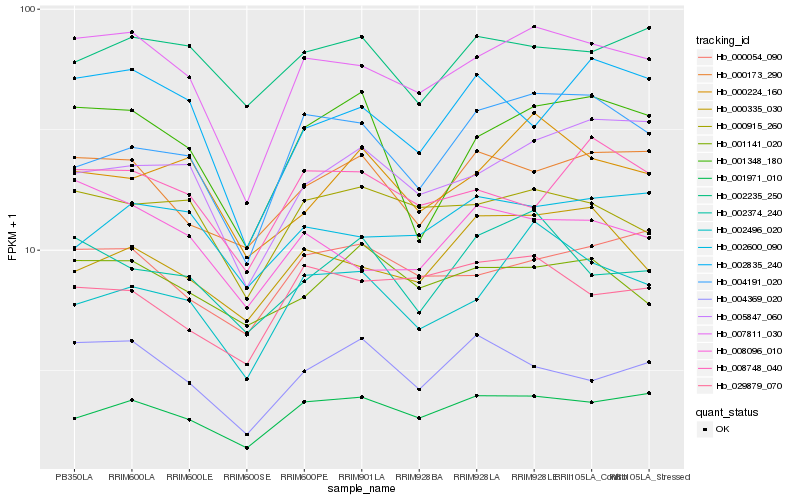
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007811_030 |
0.0 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 2 |
Hb_000915_260 |
0.086671901 |
- |
- |
PREDICTED: uncharacterized protein LOC105628514 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001971_010 |
0.0889257337 |
- |
- |
F23N19.4 [Arabidopsis thaliana] |
| 4 |
Hb_008096_010 |
0.0893545358 |
- |
- |
hypothetical protein MTR_1g045660 [Medicago truncatula] |
| 5 |
Hb_000054_090 |
0.0941844677 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 6 |
Hb_000173_290 |
0.098045539 |
- |
- |
guanylate kinase, putative [Ricinus communis] |
| 7 |
Hb_001348_180 |
0.1010565468 |
- |
- |
PREDICTED: TLC domain-containing protein 2 [Jatropha curcas] |
| 8 |
Hb_002496_020 |
0.101664542 |
- |
- |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_008748_040 |
0.1027305422 |
- |
- |
beta-hydroxyacyl-acyl carrier protein dehydratase [Hevea brasiliensis] |
| 10 |
Hb_005847_060 |
0.1042382426 |
- |
- |
hypothetical protein POPTR_0006s02920g [Populus trichocarpa] |
| 11 |
Hb_004369_020 |
0.1047081109 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g63330-like [Jatropha curcas] |
| 12 |
Hb_029879_070 |
0.1048282957 |
- |
- |
PREDICTED: phospholipase A I [Jatropha curcas] |
| 13 |
Hb_004191_020 |
0.1052275688 |
- |
- |
PREDICTED: cysteine synthase [Jatropha curcas] |
| 14 |
Hb_002835_240 |
0.1053434375 |
- |
- |
hypothetical protein PRUPE_ppa013861mg [Prunus persica] |
| 15 |
Hb_002600_090 |
0.1072429019 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_002374_240 |
0.10746379 |
- |
- |
nucellin, putative [Ricinus communis] |
| 17 |
Hb_000335_030 |
0.1085553612 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g17033 [Jatropha curcas] |
| 18 |
Hb_001141_020 |
0.1087864795 |
- |
- |
PREDICTED: stress-induced-phosphoprotein 1 [Jatropha curcas] |
| 19 |
Hb_002235_250 |
0.1093575338 |
- |
- |
PREDICTED: cystathionine gamma-synthase 1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000224_160 |
0.1093811913 |
- |
- |
PREDICTED: phenylalanine--tRNA ligase, chloroplastic/mitochondrial [Jatropha curcas] |