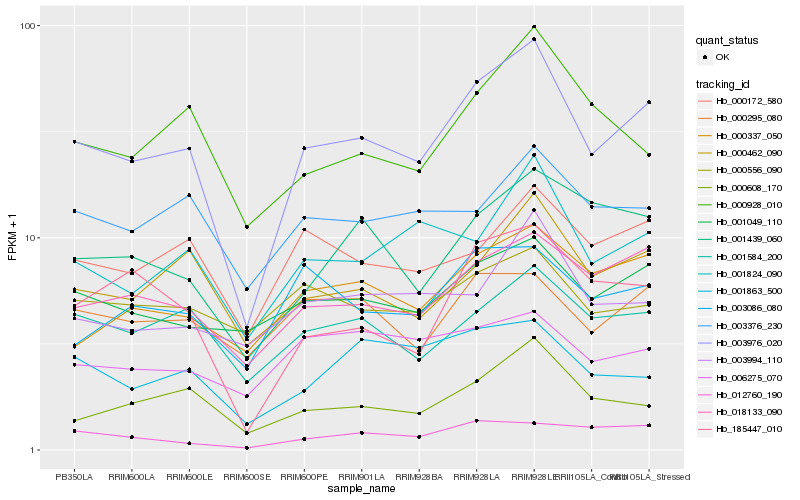
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003976_020 |
0.0 |
- |
- |
Regulator of ribonuclease activity A, putative [Ricinus communis] |
| 2 |
Hb_018133_090 |
0.1176642868 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000462_090 |
0.1247581564 |
- |
- |
PREDICTED: carotene epsilon-monooxygenase, chloroplastic [Jatropha curcas] |
| 4 |
Hb_001824_090 |
0.1288224322 |
- |
- |
- |
| 5 |
Hb_000608_170 |
0.1323759107 |
- |
- |
F23N19.4 [Arabidopsis thaliana] |
| 6 |
Hb_185447_010 |
0.1344981355 |
- |
- |
hypothetical protein JCGZ_13512 [Jatropha curcas] |
| 7 |
Hb_001584_200 |
0.1348657893 |
- |
- |
PREDICTED: Golgi apparatus membrane protein-like protein ECHIDNA [Jatropha curcas] |
| 8 |
Hb_000337_050 |
0.135677979 |
- |
- |
PREDICTED: chloroplast envelope membrane protein [Jatropha curcas] |
| 9 |
Hb_001049_110 |
0.1371690306 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 10 |
Hb_001439_060 |
0.1387921027 |
- |
- |
PREDICTED: uncharacterized protein LOC105644108 [Jatropha curcas] |
| 11 |
Hb_000172_580 |
0.1388904458 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_000928_010 |
0.1440071432 |
- |
- |
Cell division protein ftsZ, putative [Ricinus communis] |
| 13 |
Hb_003086_080 |
0.1481595414 |
- |
- |
PREDICTED: uncharacterized protein LOC105635783 isoform X3 [Jatropha curcas] |
| 14 |
Hb_012760_190 |
0.1485593515 |
- |
- |
PREDICTED: fatty acyl-CoA reductase 2 [Jatropha curcas] |
| 15 |
Hb_003994_110 |
0.1487079192 |
transcription factor |
TF Family: mTERF |
PREDICTED: uncharacterized protein LOC105646130 [Jatropha curcas] |
| 16 |
Hb_003376_230 |
0.1496811567 |
- |
- |
PREDICTED: dnaJ protein P58IPK homolog [Jatropha curcas] |
| 17 |
Hb_000295_080 |
0.150816023 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase [Jatropha curcas] |
| 18 |
Hb_001863_500 |
0.1530591677 |
- |
- |
Protein brittle-1, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_006275_070 |
0.1589946193 |
- |
- |
PREDICTED: general transcription factor IIF subunit 2 [Jatropha curcas] |
| 20 |
Hb_000556_090 |
0.1604246779 |
- |
- |
PREDICTED: uncharacterized protein LOC105643091 isoform X1 [Jatropha curcas] |