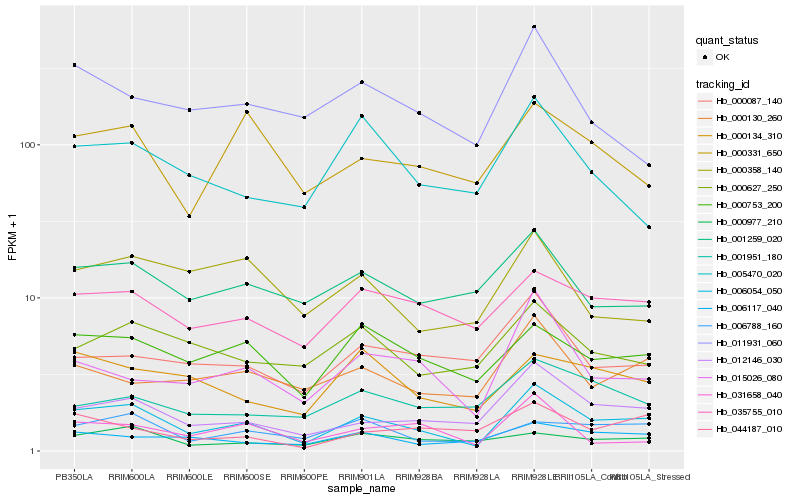
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006054_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649250, partial [Jatropha curcas] |
| 2 |
Hb_012146_030 |
0.1480458638 |
- |
- |
- |
| 3 |
Hb_015026_080 |
0.1732786264 |
- |
- |
- |
| 4 |
Hb_000753_200 |
0.1766958462 |
- |
- |
PREDICTED: putative ATP-dependent helicase hrq1 [Jatropha curcas] |
| 5 |
Hb_006788_160 |
0.1805948023 |
- |
- |
hypothetical protein POPTR_0002s04680g [Populus trichocarpa] |
| 6 |
Hb_035755_010 |
0.1810594169 |
- |
- |
unknown [Populus trichocarpa] |
| 7 |
Hb_000130_260 |
0.1850390035 |
rubber biosynthesis |
Gene Name: Dimethyallyltransferase |
PREDICTED: tRNA dimethylallyltransferase 9 isoform X1 [Jatropha curcas] |
| 8 |
Hb_006117_040 |
0.1877231162 |
- |
- |
PREDICTED: seed biotin-containing protein SBP65 [Jatropha curcas] |
| 9 |
Hb_000134_310 |
0.1890567444 |
- |
- |
PREDICTED: protein gamma response 1 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000627_250 |
0.19509186 |
- |
- |
PREDICTED: uncharacterized protein LOC105632682 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000331_650 |
0.1952737413 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase-like [Jatropha curcas] |
| 12 |
Hb_001259_020 |
0.1965106309 |
- |
- |
PREDICTED: translocase of chloroplast 90, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000087_140 |
0.1985246306 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g27270 isoform X1 [Jatropha curcas] |
| 14 |
Hb_031658_040 |
0.1995454903 |
- |
- |
- |
| 15 |
Hb_011931_060 |
0.1996241933 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP62-like [Jatropha curcas] |
| 16 |
Hb_044187_010 |
0.200232001 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 17 |
Hb_000977_210 |
0.2003163649 |
- |
- |
chromatin assembly factor 1, subunit A, putative [Ricinus communis] |
| 18 |
Hb_005470_020 |
0.2004852589 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic-like [Jatropha curcas] |
| 19 |
Hb_000358_140 |
0.2028693392 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001951_180 |
0.2032430204 |
- |
- |
PREDICTED: carboxyl-terminal-processing peptidase 1, chloroplastic isoform X2 [Jatropha curcas] |