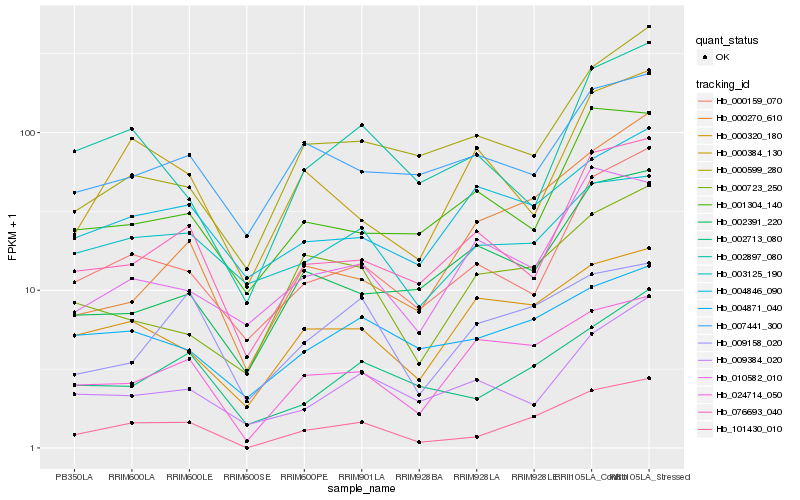
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_101430_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_002713_080 |
0.1581192989 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 3 |
Hb_024714_050 |
0.1686212638 |
- |
- |
hypothetical protein CICLE_v100188992mg, partial [Citrus clementina] |
| 4 |
Hb_000159_070 |
0.1711062541 |
- |
- |
JHL03K20.4 [Jatropha curcas] |
| 5 |
Hb_076693_040 |
0.1789806102 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5-B [Tarenaya hassleriana] |
| 6 |
Hb_000320_180 |
0.1858249653 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 7 |
Hb_000723_250 |
0.1937691023 |
- |
- |
hypothetical protein JCGZ_16520 [Jatropha curcas] |
| 8 |
Hb_009158_020 |
0.1944466985 |
- |
- |
PREDICTED: uncharacterized membrane protein At1g16860-like [Jatropha curcas] |
| 9 |
Hb_004871_040 |
0.1957107655 |
- |
- |
PREDICTED: tRNA (guanine-N(7)-)-methyltransferase [Jatropha curcas] |
| 10 |
Hb_000270_610 |
0.1960116689 |
- |
- |
PREDICTED: uncharacterized protein At5g50100, mitochondrial [Jatropha curcas] |
| 11 |
Hb_002897_080 |
0.1964465302 |
- |
- |
hypothetical protein JCGZ_03306 [Jatropha curcas] |
| 12 |
Hb_009384_020 |
0.1985125842 |
- |
- |
hypothetical protein JCGZ_24168 [Jatropha curcas] |
| 13 |
Hb_000599_280 |
0.2010414265 |
- |
- |
PREDICTED: 60S ribosomal protein L14-1 [Jatropha curcas] |
| 14 |
Hb_003125_190 |
0.2020800563 |
- |
- |
hypothetical protein POPTR_0015s10960g [Populus trichocarpa] |
| 15 |
Hb_001304_140 |
0.2027395352 |
- |
- |
PREDICTED: transmembrane protein 256 homolog [Jatropha curcas] |
| 16 |
Hb_007441_300 |
0.2049351255 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_010582_010 |
0.205023511 |
- |
- |
hypothetical protein POPTR_0011s15670g [Populus trichocarpa] |
| 18 |
Hb_004846_090 |
0.2055630302 |
transcription factor |
TF Family: HMG |
HMG-box DNA-binding family protein isoform 1 [Theobroma cacao] |
| 19 |
Hb_002391_220 |
0.2064359981 |
- |
- |
PREDICTED: UPF0548 protein At2g17695 [Jatropha curcas] |
| 20 |
Hb_000384_130 |
0.2070835574 |
- |
- |
uridylate kinase plant, putative [Ricinus communis] |