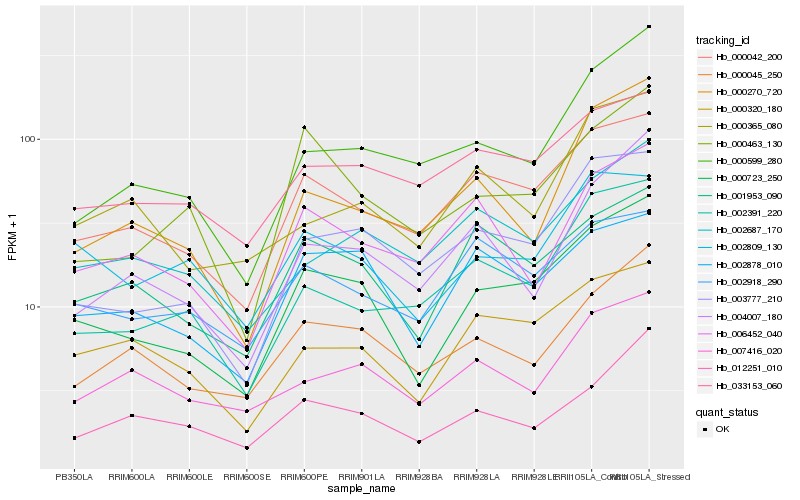
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000723_250 |
0.0 |
- |
- |
hypothetical protein JCGZ_16520 [Jatropha curcas] |
| 2 |
Hb_000042_200 |
0.107793049 |
- |
- |
PREDICTED: uncharacterized protein LOC100252136 isoform X1 [Vitis vinifera] |
| 3 |
Hb_003777_210 |
0.11616616 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 4 |
Hb_001953_090 |
0.1229470281 |
- |
- |
PREDICTED: membrane-anchored ubiquitin-fold protein 3 [Jatropha curcas] |
| 5 |
Hb_000320_180 |
0.1264294701 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 6 |
Hb_000045_250 |
0.1284359493 |
- |
- |
PREDICTED: uncharacterized protein LOC105646572 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002878_010 |
0.1302804209 |
- |
- |
- |
| 8 |
Hb_000463_130 |
0.1387046924 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_002809_130 |
0.1402555146 |
- |
- |
mads box protein, putative [Ricinus communis] |
| 10 |
Hb_033153_060 |
0.1450191843 |
- |
- |
Transcription elongation factor, putative [Ricinus communis] |
| 11 |
Hb_012251_010 |
0.1452400732 |
- |
- |
- |
| 12 |
Hb_000599_280 |
0.1453357908 |
- |
- |
PREDICTED: 60S ribosomal protein L14-1 [Jatropha curcas] |
| 13 |
Hb_002687_170 |
0.1455312447 |
- |
- |
PREDICTED: G patch domain and ankyrin repeat-containing protein 1 homolog [Jatropha curcas] |
| 14 |
Hb_002391_220 |
0.1469559805 |
- |
- |
PREDICTED: UPF0548 protein At2g17695 [Jatropha curcas] |
| 15 |
Hb_004007_180 |
0.1499355187 |
- |
- |
PREDICTED: uncharacterized protein LOC105639838 [Jatropha curcas] |
| 16 |
Hb_007416_020 |
0.1503306308 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Cucumis sativus] |
| 17 |
Hb_000270_720 |
0.1503348489 |
- |
- |
hypothetical protein POPTR_0015s08460g [Populus trichocarpa] |
| 18 |
Hb_002918_290 |
0.1520929023 |
- |
- |
PREDICTED: uncharacterized protein LOC105649583 isoform X1 [Jatropha curcas] |
| 19 |
Hb_000365_080 |
0.1534088702 |
- |
- |
PREDICTED: uncharacterized protein LOC105649037 [Jatropha curcas] |
| 20 |
Hb_006452_040 |
0.1536876329 |
- |
- |
PREDICTED: signal recognition particle 9 kDa protein [Jatropha curcas] |