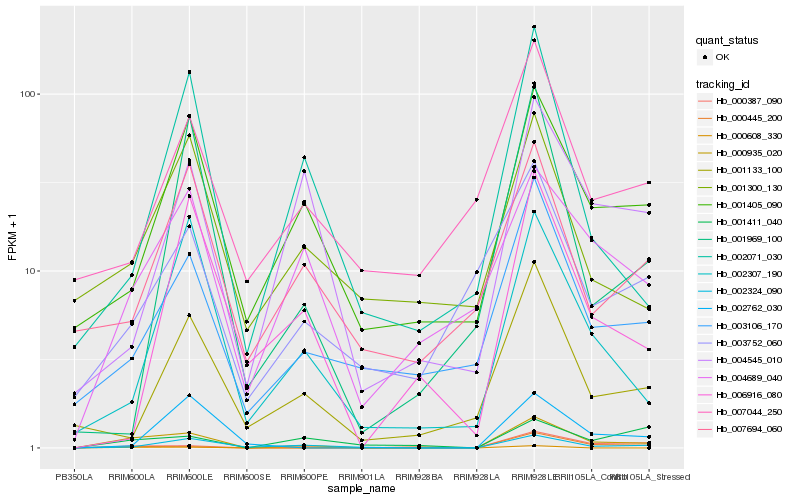
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000935_020 |
0.0 |
- |
- |
putative non-LTR retroelement reverse transcriptase [Arabidopsis thaliana] |
| 2 |
Hb_002324_090 |
0.2584654983 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 3 |
Hb_003106_170 |
0.278764633 |
- |
- |
Endonuclease III, putative [Ricinus communis] |
| 4 |
Hb_000387_090 |
0.2820836149 |
- |
- |
PREDICTED: uncharacterized protein LOC104115978 [Nicotiana tomentosiformis] |
| 5 |
Hb_001405_090 |
0.2856406189 |
- |
- |
PREDICTED: photosynthetic NDH subunit of subcomplex B 5, chloroplastic [Jatropha curcas] |
| 6 |
Hb_004689_040 |
0.2899531357 |
- |
- |
PREDICTED: uncharacterized protein LOC105648084 [Jatropha curcas] |
| 7 |
Hb_002307_190 |
0.2954856286 |
- |
- |
PREDICTED: putative ion channel POLLUX-like 2 [Jatropha curcas] |
| 8 |
Hb_000445_200 |
0.2986643788 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 9 |
Hb_001133_100 |
0.3035092462 |
- |
- |
PREDICTED: oligopeptide transporter 6-like [Jatropha curcas] |
| 10 |
Hb_000608_330 |
0.3038622446 |
- |
- |
Zeamatin precursor, putative [Ricinus communis] |
| 11 |
Hb_002071_030 |
0.3067988985 |
- |
- |
PREDICTED: ACT domain-containing protein ACR11-like isoform X1 [Gossypium raimondii] |
| 12 |
Hb_002762_030 |
0.307726364 |
- |
- |
hypothetical protein POPTR_0019s12370g, partial [Populus trichocarpa] |
| 13 |
Hb_004545_010 |
0.3106593474 |
- |
- |
acyl carrier protein [Ricinus communis] |
| 14 |
Hb_003752_060 |
0.3126443426 |
- |
- |
PREDICTED: epoxide hydrolase 3 [Jatropha curcas] |
| 15 |
Hb_001411_040 |
0.3172692541 |
- |
- |
hypothetical protein VITISV_010644 [Vitis vinifera] |
| 16 |
Hb_007694_060 |
0.3203370114 |
- |
- |
PREDICTED: 50S ribosomal protein L35, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001300_130 |
0.3206140418 |
- |
- |
PREDICTED: protein TRANSPORT INHIBITOR RESPONSE 1 [Jatropha curcas] |
| 18 |
Hb_001969_100 |
0.321336737 |
- |
- |
signal transducer, putative [Ricinus communis] |
| 19 |
Hb_007044_250 |
0.3214317005 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_006916_080 |
0.3216063007 |
- |
- |
PREDICTED: uncharacterized protein LOC105646751 [Jatropha curcas] |