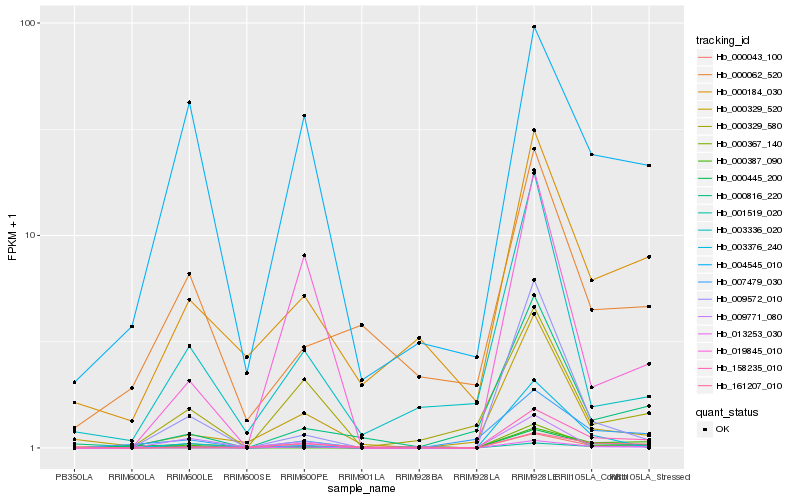
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_013253_030 |
0.0 |
- |
- |
PREDICTED: protein argonaute 16 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000043_100 |
0.1875344697 |
- |
- |
PREDICTED: uncharacterized protein LOC105797583 [Gossypium raimondii] |
| 3 |
Hb_158235_010 |
0.2115744634 |
- |
- |
PREDICTED: uncharacterized protein LOC104800023, partial [Tarenaya hassleriana] |
| 4 |
Hb_000445_200 |
0.2349048966 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 5 |
Hb_009572_010 |
0.2403192329 |
- |
- |
PREDICTED: MLP-like protein 423 [Jatropha curcas] |
| 6 |
Hb_000816_220 |
0.2429536415 |
- |
- |
PREDICTED: CBL-interacting protein kinase 18-like [Jatropha curcas] |
| 7 |
Hb_000329_580 |
0.2441778277 |
- |
- |
malate dehydrogenase, putative [Ricinus communis] |
| 8 |
Hb_004545_010 |
0.2456622732 |
- |
- |
acyl carrier protein [Ricinus communis] |
| 9 |
Hb_007479_030 |
0.2472528886 |
- |
- |
PREDICTED: asparagine synthetase [glutamine-hydrolyzing] 3 [Jatropha curcas] |
| 10 |
Hb_000184_030 |
0.2517733633 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 11 |
Hb_003376_240 |
0.254746585 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_019845_010 |
0.2561107658 |
- |
- |
protease inhibitor protein [Hevea brasiliensis] |
| 13 |
Hb_000329_520 |
0.2592562124 |
- |
- |
PREDICTED: metalloendoproteinase 1 [Jatropha curcas] |
| 14 |
Hb_001519_020 |
0.2663549247 |
- |
- |
hypothetical protein JCGZ_09913 [Jatropha curcas] |
| 15 |
Hb_000367_140 |
0.266355528 |
- |
- |
S-locus-specific glycoprotein S6 precursor, putative [Ricinus communis] |
| 16 |
Hb_000387_090 |
0.2757128878 |
- |
- |
PREDICTED: uncharacterized protein LOC104115978 [Nicotiana tomentosiformis] |
| 17 |
Hb_161207_010 |
0.2824103469 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 18 |
Hb_003336_020 |
0.2827919599 |
- |
- |
PREDICTED: probable sodium/metabolite cotransporter BASS3, chloroplastic [Vitis vinifera] |
| 19 |
Hb_009771_080 |
0.2860001137 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 20 |
Hb_000062_520 |
0.2876999869 |
- |
- |
conserved hypothetical protein [Ricinus communis] |