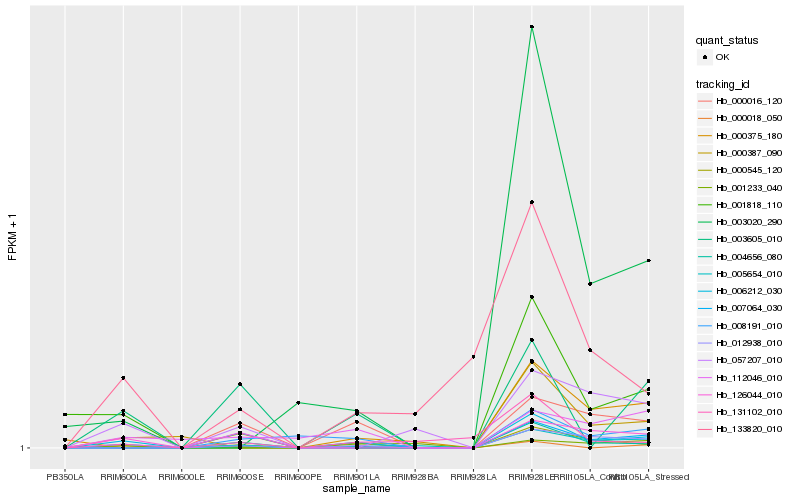
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006212_030 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 2 |
Hb_003020_290 |
0.2825950959 |
- |
- |
hypothetical protein CICLE_v10029104mg [Citrus clementina] |
| 3 |
Hb_000018_050 |
0.2854052426 |
- |
- |
PREDICTED: uncharacterized protein LOC104899363 [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_012938_010 |
0.2878367489 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 5 |
Hb_005654_010 |
0.3151675241 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_000545_120 |
0.316191796 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 7 |
Hb_000387_090 |
0.3174429233 |
- |
- |
PREDICTED: uncharacterized protein LOC104115978 [Nicotiana tomentosiformis] |
| 8 |
Hb_000375_180 |
0.3211328797 |
- |
- |
PREDICTED: uncharacterized protein LOC104593656 [Nelumbo nucifera] |
| 9 |
Hb_007064_030 |
0.324483586 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 10 |
Hb_126044_010 |
0.3245444452 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 11 |
Hb_133820_010 |
0.3290815549 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 12 |
Hb_001818_110 |
0.3303420982 |
- |
- |
- |
| 13 |
Hb_003605_010 |
0.3314478011 |
- |
- |
- |
| 14 |
Hb_112046_010 |
0.3316130632 |
- |
- |
PREDICTED: uncharacterized protein LOC105772042 [Gossypium raimondii] |
| 15 |
Hb_000016_120 |
0.3405426091 |
- |
- |
PREDICTED: uncharacterized protein LOC104610208 [Nelumbo nucifera] |
| 16 |
Hb_001233_040 |
0.3420742679 |
- |
- |
PREDICTED: uncharacterized protein LOC105775263 [Gossypium raimondii] |
| 17 |
Hb_057207_010 |
0.3439683851 |
- |
- |
hypothetical protein F383_17010 [Gossypium arboreum] |
| 18 |
Hb_131102_010 |
0.3475698591 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 19 |
Hb_008191_010 |
0.353107282 |
- |
- |
- |
| 20 |
Hb_004656_080 |
0.3531463752 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |