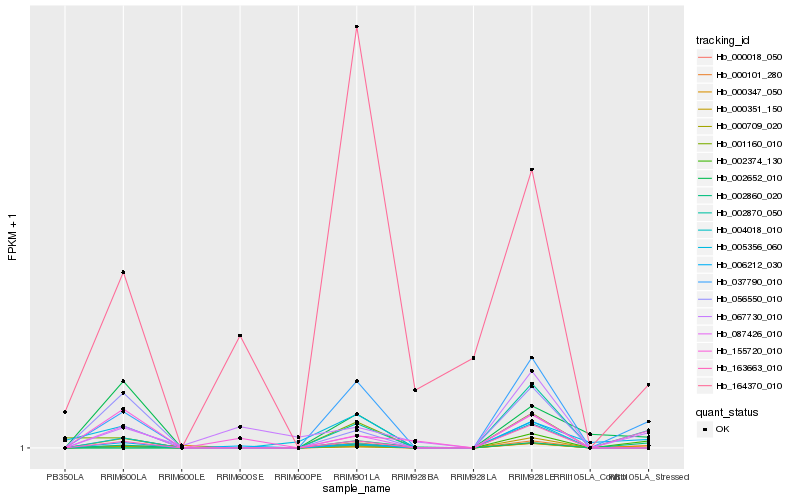
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_037790_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 2 |
Hb_000018_050 |
0.2174980537 |
- |
- |
PREDICTED: uncharacterized protein LOC104899363 [Beta vulgaris subsp. vulgaris] |
| 3 |
Hb_002860_020 |
0.2393550717 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 4 |
Hb_000101_280 |
0.2429266325 |
- |
- |
hypothetical protein POPTR_0018s02450g [Populus trichocarpa] |
| 5 |
Hb_163663_010 |
0.2843788013 |
- |
- |
PREDICTED: uncharacterized protein LOC105629934 [Jatropha curcas] |
| 6 |
Hb_087426_010 |
0.3003352325 |
- |
- |
PREDICTED: uncharacterized protein LOC105803348 [Gossypium raimondii] |
| 7 |
Hb_000709_020 |
0.3045760771 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 8 |
Hb_002870_050 |
0.3143494449 |
- |
- |
- |
| 9 |
Hb_002652_010 |
0.3356946355 |
- |
- |
- |
| 10 |
Hb_001160_010 |
0.3447212616 |
- |
- |
- |
| 11 |
Hb_000347_050 |
0.3550386906 |
- |
- |
PREDICTED: ABC transporter B family member 3-like [Jatropha curcas] |
| 12 |
Hb_056550_010 |
0.3574908557 |
- |
- |
PREDICTED: uncharacterized protein LOC104112577 [Nicotiana tomentosiformis] |
| 13 |
Hb_006212_030 |
0.3593232902 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 14 |
Hb_002374_130 |
0.359978787 |
- |
- |
PREDICTED: metal transporter Nramp5-like [Populus euphratica] |
| 15 |
Hb_000351_150 |
0.3660171794 |
- |
- |
Wall-associated kinase 2, putative [Theobroma cacao] |
| 16 |
Hb_164370_010 |
0.3683088584 |
- |
- |
- |
| 17 |
Hb_004018_010 |
0.3692174916 |
- |
- |
PREDICTED: uncharacterized protein LOC105784835 [Gossypium raimondii] |
| 18 |
Hb_005356_060 |
0.3781467808 |
- |
- |
hypothetical protein POPTR_0013s14620g, partial [Populus trichocarpa] |
| 19 |
Hb_155720_010 |
0.3793160784 |
- |
- |
PREDICTED: uncharacterized protein LOC104115978 [Nicotiana tomentosiformis] |
| 20 |
Hb_067730_010 |
0.3834806609 |
- |
- |
hypothetical protein OsJ_21419 [Oryza sativa Japonica Group] |