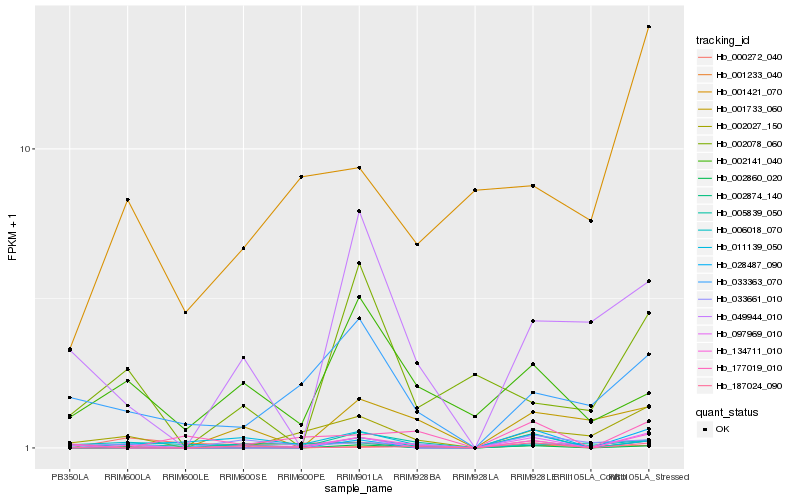
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_134711_010 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_D00316 [Eucalyptus grandis] |
| 2 |
Hb_002860_020 |
0.2634045862 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 3 |
Hb_002027_150 |
0.3158006621 |
- |
- |
- |
| 4 |
Hb_011139_050 |
0.3462582543 |
- |
- |
nicotinate phosphoribosyltransferase family protein [Populus trichocarpa] |
| 5 |
Hb_001233_040 |
0.3628810626 |
- |
- |
PREDICTED: uncharacterized protein LOC105775263 [Gossypium raimondii] |
| 6 |
Hb_187024_090 |
0.3664986323 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_033363_070 |
0.3704075997 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: methionine aminopeptidase 1B, chloroplastic, partial [Populus euphratica] |
| 8 |
Hb_001733_060 |
0.3752117819 |
- |
- |
PREDICTED: uncharacterized protein LOC104431099 [Eucalyptus grandis] |
| 9 |
Hb_001421_070 |
0.3794867551 |
- |
- |
PREDICTED: ribose-phosphate pyrophosphokinase 1 [Jatropha curcas] |
| 10 |
Hb_002874_140 |
0.3856231433 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 11 homolog [Prunus mume] |
| 11 |
Hb_028487_090 |
0.3857368138 |
- |
- |
- |
| 12 |
Hb_006018_070 |
0.3864239782 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 13 |
Hb_049944_010 |
0.3864795284 |
- |
- |
transducin family protein [Populus trichocarpa] |
| 14 |
Hb_097969_010 |
0.3871482108 |
- |
- |
F-box/LRR-repeat protein, putative [Ricinus communis] |
| 15 |
Hb_002141_040 |
0.3878359198 |
- |
- |
- |
| 16 |
Hb_033661_010 |
0.3891491202 |
- |
- |
PREDICTED: uncharacterized protein LOC105852393, partial [Cicer arietinum] |
| 17 |
Hb_177019_010 |
0.3895708786 |
- |
- |
Uncharacterized protein TCM_004012 [Theobroma cacao] |
| 18 |
Hb_005839_050 |
0.3900836879 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 19 |
Hb_002078_060 |
0.3907901609 |
- |
- |
hypothetical protein JCGZ_17943 [Jatropha curcas] |
| 20 |
Hb_000272_040 |
0.3912696786 |
- |
- |
- |