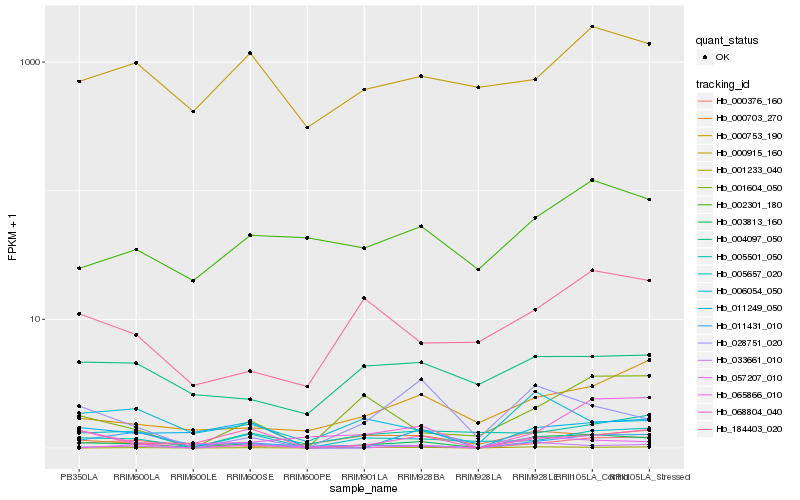
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003813_160 |
0.0 |
- |
- |
- |
| 2 |
Hb_001233_040 |
0.2705337712 |
- |
- |
PREDICTED: uncharacterized protein LOC105775263 [Gossypium raimondii] |
| 3 |
Hb_001604_050 |
0.2707014785 |
- |
- |
PREDICTED: uncharacterized protein LOC105639208 [Jatropha curcas] |
| 4 |
Hb_033661_010 |
0.2754734314 |
- |
- |
PREDICTED: uncharacterized protein LOC105852393, partial [Cicer arietinum] |
| 5 |
Hb_184403_020 |
0.2807876863 |
- |
- |
PREDICTED: pre-rRNA-processing protein ESF2 [Vitis vinifera] |
| 6 |
Hb_000753_190 |
0.2822831271 |
- |
- |
PREDICTED: D-aminoacyl-tRNA deacylase [Populus euphratica] |
| 7 |
Hb_011249_050 |
0.2859686507 |
- |
- |
- |
| 8 |
Hb_028751_020 |
0.2865572585 |
- |
- |
PREDICTED: SCY1-like protein 2 [Jatropha curcas] |
| 9 |
Hb_068804_040 |
0.2868780741 |
- |
- |
- |
| 10 |
Hb_002301_180 |
0.2905234268 |
- |
- |
PREDICTED: protein yippee-like At5g53940 [Jatropha curcas] |
| 11 |
Hb_065866_010 |
0.2916425849 |
- |
- |
PREDICTED: hydroxyacyl-thioester dehydratase type 2, mitochondrial [Fragaria vesca subsp. vesca] |
| 12 |
Hb_000703_270 |
0.291702584 |
- |
- |
hypothetical protein B456_012G039600 [Gossypium raimondii] |
| 13 |
Hb_000376_160 |
0.2956575177 |
- |
- |
- |
| 14 |
Hb_057207_010 |
0.2956856332 |
- |
- |
hypothetical protein F383_17010 [Gossypium arboreum] |
| 15 |
Hb_011431_010 |
0.2988641845 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 16 |
Hb_005501_050 |
0.300348058 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 17 |
Hb_005657_020 |
0.3007029729 |
- |
- |
- |
| 18 |
Hb_006054_050 |
0.3008597402 |
- |
- |
PREDICTED: uncharacterized protein LOC105649250, partial [Jatropha curcas] |
| 19 |
Hb_000915_160 |
0.3049263306 |
- |
- |
PREDICTED: histone H1 [Jatropha curcas] |
| 20 |
Hb_004097_050 |
0.3054648974 |
- |
- |
PREDICTED: squalene monooxygenase-like [Jatropha curcas] |