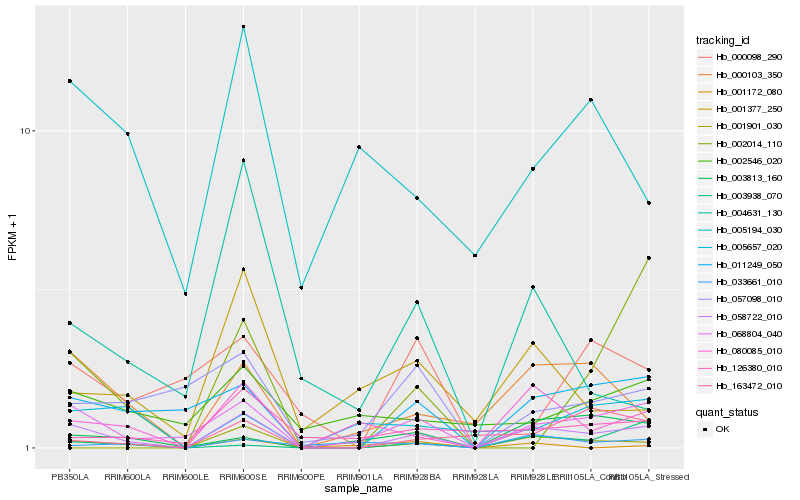
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_058722_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_068804_040 |
0.2098998864 |
- |
- |
- |
| 3 |
Hb_011249_050 |
0.2591518367 |
- |
- |
- |
| 4 |
Hb_000103_350 |
0.2814852788 |
- |
- |
- |
| 5 |
Hb_080085_010 |
0.282598773 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 6 |
Hb_003813_160 |
0.3278142066 |
- |
- |
- |
| 7 |
Hb_057098_010 |
0.3460627653 |
- |
- |
- |
| 8 |
Hb_163472_010 |
0.3464625668 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 9 |
Hb_002014_110 |
0.3512495117 |
- |
- |
- |
| 10 |
Hb_001377_250 |
0.3540331241 |
- |
- |
dynamin, putative [Ricinus communis] |
| 11 |
Hb_033661_010 |
0.3566885961 |
- |
- |
PREDICTED: uncharacterized protein LOC105852393, partial [Cicer arietinum] |
| 12 |
Hb_003938_070 |
0.3607575637 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 13 |
Hb_126380_010 |
0.3612226485 |
desease resistance |
Gene Name: NB-ARC |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_005194_030 |
0.3617437541 |
- |
- |
PREDICTED: putative formamidase C869.04 isoform X1 [Jatropha curcas] |
| 15 |
Hb_004631_130 |
0.3623641529 |
- |
- |
helicase domain-containing family protein [Populus trichocarpa] |
| 16 |
Hb_002546_020 |
0.3628958273 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase SD1-1 [Malus domestica] |
| 17 |
Hb_001172_080 |
0.3631899004 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 18 |
Hb_001901_030 |
0.3664426735 |
- |
- |
- |
| 19 |
Hb_005657_020 |
0.3667325875 |
- |
- |
- |
| 20 |
Hb_000098_290 |
0.3675214088 |
- |
- |
- |