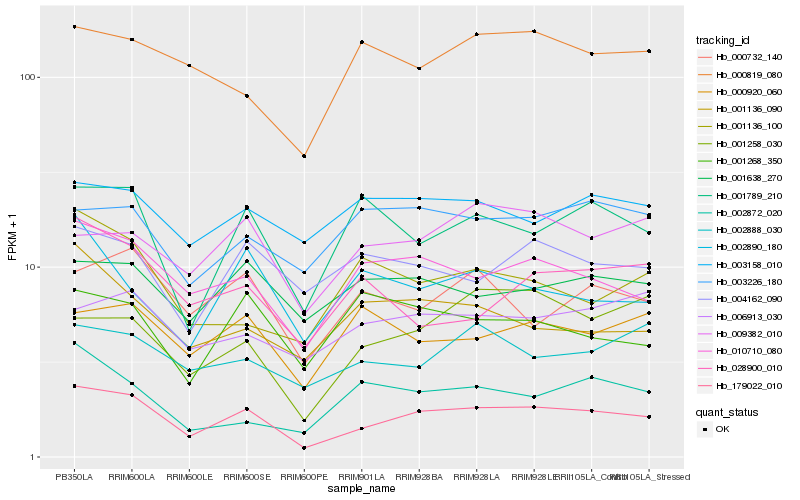
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_179022_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 2 |
Hb_010710_080 |
0.1159088136 |
- |
- |
- |
| 3 |
Hb_001789_210 |
0.1402932168 |
- |
- |
PREDICTED: methyltransferase-like protein 1 isoform X2 [Jatropha curcas] |
| 4 |
Hb_001638_270 |
0.1460656069 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 8 homolog isoform X2 [Jatropha curcas] |
| 5 |
Hb_000732_140 |
0.1463734915 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase abkC [Jatropha curcas] |
| 6 |
Hb_004162_090 |
0.1471690254 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor DIVARICATA [Jatropha curcas] |
| 7 |
Hb_001136_090 |
0.1522652985 |
- |
- |
Protein PNS1, putative [Ricinus communis] |
| 8 |
Hb_009382_010 |
0.1525979162 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_002888_030 |
0.1547722853 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1 [Jatropha curcas] |
| 10 |
Hb_001258_030 |
0.154969285 |
- |
- |
tRNA-dihydrouridine synthase, putative [Ricinus communis] |
| 11 |
Hb_006913_030 |
0.1553255116 |
- |
- |
PREDICTED: quinone oxidoreductase-like protein 2 homolog [Jatropha curcas] |
| 12 |
Hb_003158_010 |
0.1553369754 |
- |
- |
hypothetical protein JCGZ_17361 [Jatropha curcas] |
| 13 |
Hb_002890_180 |
0.1555049509 |
- |
- |
PREDICTED: squamous cell carcinoma antigen recognized by T-cells 3 [Jatropha curcas] |
| 14 |
Hb_003226_180 |
0.15616531 |
- |
- |
PREDICTED: uncharacterized protein C57A7.06 [Jatropha curcas] |
| 15 |
Hb_000819_080 |
0.1568422341 |
- |
- |
Plastid-lipid-associated protein, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_000920_060 |
0.156983627 |
- |
- |
PREDICTED: intron-binding protein aquarius [Jatropha curcas] |
| 17 |
Hb_028900_010 |
0.1593853264 |
- |
- |
PREDICTED: uncharacterized protein LOC105631404 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001136_100 |
0.1603304881 |
- |
- |
hypothetical protein MTR_085s0013 [Medicago truncatula] |
| 19 |
Hb_002872_020 |
0.1603381208 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 20 |
Hb_001268_350 |
0.1605471823 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105115846 [Populus euphratica] |