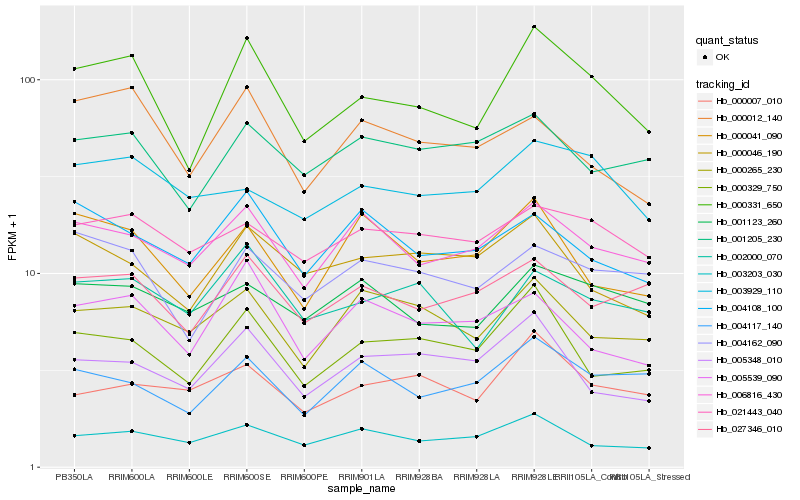
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000331_650 |
0.0 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase-like [Jatropha curcas] |
| 2 |
Hb_000329_750 |
0.1138209066 |
- |
- |
PREDICTED: uncharacterized protein LOC105643205 [Jatropha curcas] |
| 3 |
Hb_004162_090 |
0.1195054922 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor DIVARICATA [Jatropha curcas] |
| 4 |
Hb_002000_070 |
0.1222599778 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: 5-formyltetrahydrofolate cyclo-ligase-like protein COG0212 [Jatropha curcas] |
| 5 |
Hb_027346_010 |
0.1229129061 |
- |
- |
- |
| 6 |
Hb_006816_430 |
0.1233115196 |
- |
- |
PREDICTED: topless-related protein 3 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004117_140 |
0.1263006847 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At3g16710, mitochondrial [Populus euphratica] |
| 8 |
Hb_005539_090 |
0.1298512607 |
transcription factor |
TF Family: PHD |
PREDICTED: methyl-CpG-binding domain-containing protein 9 isoform X1 [Jatropha curcas] |
| 9 |
Hb_003203_030 |
0.1307299567 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g13600-like [Jatropha curcas] |
| 10 |
Hb_000012_140 |
0.1318603122 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001205_230 |
0.1321619562 |
- |
- |
PREDICTED: acidic leucine-rich nuclear phosphoprotein 32-related protein [Jatropha curcas] |
| 12 |
Hb_005348_010 |
0.1332210639 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000046_190 |
0.1336480013 |
- |
- |
PREDICTED: uncharacterized protein LOC105647118 [Jatropha curcas] |
| 14 |
Hb_000041_090 |
0.133731643 |
- |
- |
PREDICTED: uncharacterized protein LOC105641452 [Jatropha curcas] |
| 15 |
Hb_021443_040 |
0.1358317611 |
- |
- |
acc synthase, putative [Ricinus communis] |
| 16 |
Hb_001123_260 |
0.1366963149 |
- |
- |
PREDICTED: ankyrin repeat and SAM domain-containing protein 3-like [Jatropha curcas] |
| 17 |
Hb_000265_230 |
0.1372876525 |
transcription factor |
TF Family: HB |
PREDICTED: uncharacterized protein LOC105643603 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000007_010 |
0.138289252 |
- |
- |
PREDICTED: protein PHYLLO, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003929_110 |
0.1397604319 |
- |
- |
PREDICTED: probable GTP diphosphokinase CRSH, chloroplastic isoform X1 [Jatropha curcas] |
| 20 |
Hb_004108_100 |
0.1402585746 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 33B isoform X2 [Jatropha curcas] |