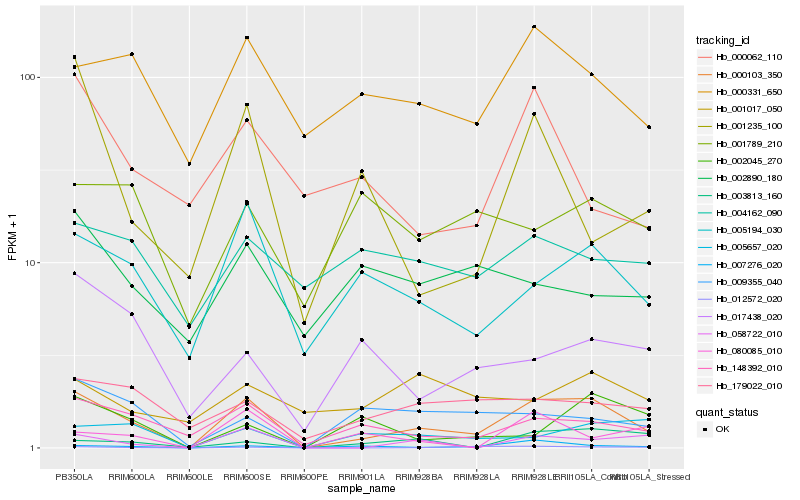
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000103_350 |
0.0 |
- |
- |
- |
| 2 |
Hb_148392_010 |
0.2389926737 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 3 |
Hb_005194_030 |
0.2511770835 |
- |
- |
PREDICTED: putative formamidase C869.04 isoform X1 [Jatropha curcas] |
| 4 |
Hb_012572_020 |
0.2739589653 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 5 |
Hb_000331_650 |
0.2749790305 |
- |
- |
PREDICTED: peptide methionine sulfoxide reductase-like [Jatropha curcas] |
| 6 |
Hb_179022_010 |
0.2791071195 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 7 |
Hb_058722_010 |
0.2814852788 |
- |
- |
- |
| 8 |
Hb_009355_040 |
0.284190735 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 9 |
Hb_007276_020 |
0.2864205306 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 10 |
Hb_005657_020 |
0.2919548396 |
- |
- |
- |
| 11 |
Hb_017438_020 |
0.2990656047 |
- |
- |
- |
| 12 |
Hb_002045_270 |
0.3038811077 |
- |
- |
hypothetical protein JCGZ_21864 [Jatropha curcas] |
| 13 |
Hb_001235_100 |
0.3094764243 |
- |
- |
heat-shock protein, putative [Ricinus communis] |
| 14 |
Hb_000062_110 |
0.3113976711 |
transcription factor |
TF Family: MYB |
hypothetical protein JCGZ_17894 [Jatropha curcas] |
| 15 |
Hb_004162_090 |
0.3131578924 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor DIVARICATA [Jatropha curcas] |
| 16 |
Hb_001789_210 |
0.3153157282 |
- |
- |
PREDICTED: methyltransferase-like protein 1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_002890_180 |
0.3159099963 |
- |
- |
PREDICTED: squamous cell carcinoma antigen recognized by T-cells 3 [Jatropha curcas] |
| 18 |
Hb_003813_160 |
0.3177196068 |
- |
- |
- |
| 19 |
Hb_001017_050 |
0.3205489535 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 20 |
Hb_080085_010 |
0.3206734607 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |