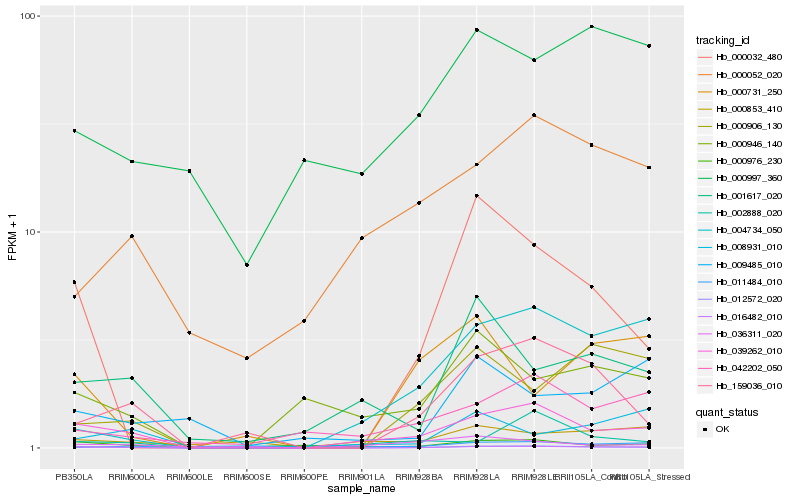
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_016482_010 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_039262_010 |
0.1580027913 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 3 |
Hb_159036_010 |
0.2249145692 |
- |
- |
- |
| 4 |
Hb_012572_020 |
0.2482591367 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 5 |
Hb_000032_480 |
0.2602063258 |
- |
- |
hypothetical protein glysoja_043543 [Glycine soja] |
| 6 |
Hb_000906_130 |
0.263706043 |
- |
- |
- |
| 7 |
Hb_001617_020 |
0.2853252568 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 8 |
Hb_000976_230 |
0.2921977731 |
- |
- |
PREDICTED: probable terpene synthase 4 [Jatropha curcas] |
| 9 |
Hb_000946_140 |
0.2952052454 |
- |
- |
- |
| 10 |
Hb_042202_050 |
0.298929273 |
- |
- |
TPA: hypothetical protein ZEAMMB73_683745 [Zea mays] |
| 11 |
Hb_004734_050 |
0.3009132319 |
- |
- |
- |
| 12 |
Hb_000052_020 |
0.3094570755 |
desease resistance |
Gene Name: ABC_tran |
PREDICTED: LOW QUALITY PROTEIN: ABC transporter G family member 2-like [Jatropha curcas] |
| 13 |
Hb_009485_010 |
0.3113987906 |
- |
- |
PREDICTED: uncharacterized protein LOC105641071 [Jatropha curcas] |
| 14 |
Hb_008931_010 |
0.313163013 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 15 |
Hb_011484_010 |
0.3142724098 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 16 |
Hb_000731_250 |
0.3148463031 |
- |
- |
- |
| 17 |
Hb_036311_020 |
0.316031677 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 18 |
Hb_002888_020 |
0.3167286577 |
- |
- |
- |
| 19 |
Hb_000853_410 |
0.3183971876 |
- |
- |
- |
| 20 |
Hb_000997_360 |
0.3198077682 |
- |
- |
hypothetical protein AALP_AAs70636U000100 [Arabis alpina] |