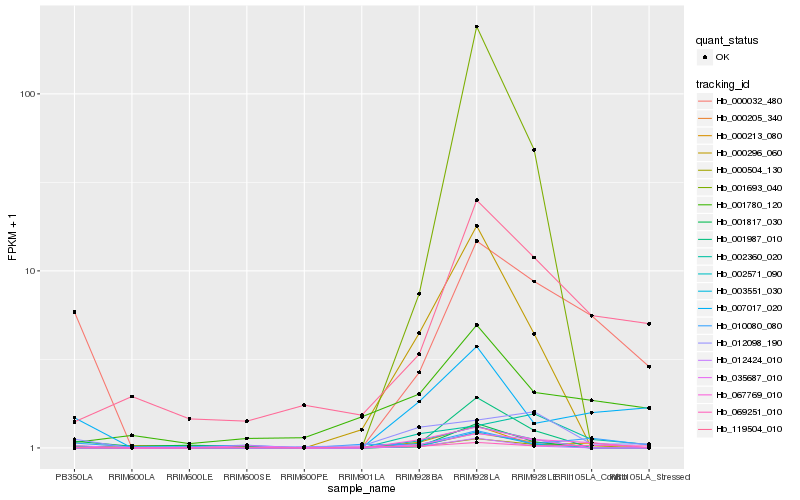
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000504_130 |
0.0 |
- |
- |
GPI transamidase component family protein [Populus trichocarpa] |
| 2 |
Hb_000032_480 |
0.1951115549 |
- |
- |
hypothetical protein glysoja_043543 [Glycine soja] |
| 3 |
Hb_067769_010 |
0.2619747251 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 4 |
Hb_007017_020 |
0.3064200498 |
- |
- |
hypothetical protein JCGZ_21740 [Jatropha curcas] |
| 5 |
Hb_035687_010 |
0.3242962987 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 6 |
Hb_119504_010 |
0.3319149025 |
- |
- |
hypothetical protein JCGZ_15304 [Jatropha curcas] |
| 7 |
Hb_002360_020 |
0.344050671 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 8 |
Hb_000205_340 |
0.3448879618 |
- |
- |
PREDICTED: putative B3 domain-containing protein At2g27410 [Jatropha curcas] |
| 9 |
Hb_000213_080 |
0.3487990146 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IX.1-like [Vitis vinifera] |
| 10 |
Hb_012098_190 |
0.3528396121 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 11 |
Hb_001987_010 |
0.3553211286 |
- |
- |
- |
| 12 |
Hb_003551_030 |
0.3567729332 |
- |
- |
hypothetical protein POPTR_0016s03850g [Populus trichocarpa] |
| 13 |
Hb_002571_090 |
0.359404364 |
- |
- |
- |
| 14 |
Hb_000296_060 |
0.3692878309 |
- |
- |
- |
| 15 |
Hb_001817_030 |
0.3708411178 |
- |
- |
PREDICTED: putative CCR4-associated factor 1 homolog 8 [Jatropha curcas] |
| 16 |
Hb_069251_010 |
0.3759540952 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 17 |
Hb_012424_010 |
0.3760619099 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 18 |
Hb_001780_120 |
0.3769327355 |
- |
- |
PREDICTED: uncharacterized protein LOC105634429 [Jatropha curcas] |
| 19 |
Hb_001693_040 |
0.3808430243 |
- |
- |
- |
| 20 |
Hb_010080_080 |
0.3816769318 |
- |
- |
conserved hypothetical protein [Ricinus communis] |