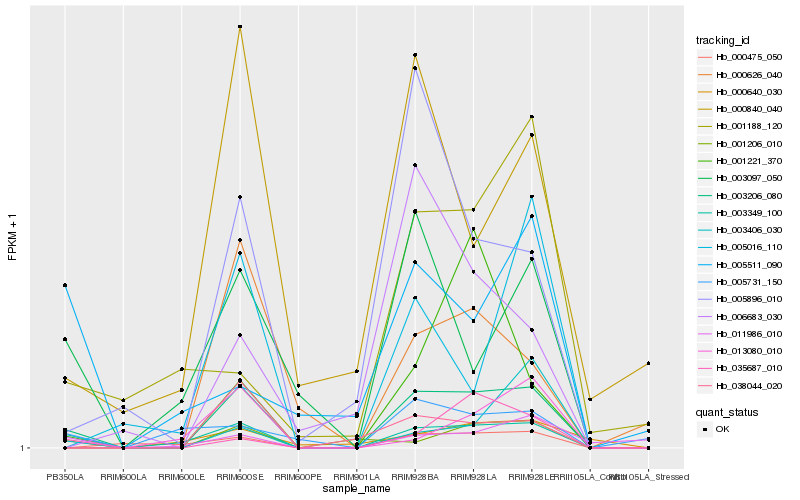
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003206_080 |
0.0 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox-leucine zipper protein HAT22-like [Jatropha curcas] |
| 2 |
Hb_003349_100 |
0.1757049508 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_005896_010 |
0.2821426841 |
- |
- |
- |
| 4 |
Hb_003097_050 |
0.2894699899 |
- |
- |
hypothetical protein POPTR_0007s07150g [Populus trichocarpa] |
| 5 |
Hb_035687_010 |
0.29405008 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 6 |
Hb_038044_020 |
0.2985129738 |
- |
- |
- |
| 7 |
Hb_001221_370 |
0.3081977599 |
- |
- |
hypothetical protein MIMGU_mgv1a020064mg, partial [Erythranthe guttata] |
| 8 |
Hb_000840_040 |
0.3143661605 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 9 |
Hb_000475_050 |
0.3158065343 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 10 |
Hb_000626_040 |
0.3217363957 |
- |
- |
PREDICTED: TATA-box-binding protein 1-like [Jatropha curcas] |
| 11 |
Hb_003406_030 |
0.3222052714 |
- |
- |
PREDICTED: uncharacterized protein LOC104774100 [Camelina sativa] |
| 12 |
Hb_005016_110 |
0.3244103754 |
- |
- |
- |
| 13 |
Hb_011986_010 |
0.3314338619 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 14 |
Hb_000640_030 |
0.3315858187 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 15 |
Hb_005731_150 |
0.3439738493 |
- |
- |
PREDICTED: uncharacterized protein LOC105630360 [Jatropha curcas] |
| 16 |
Hb_006683_030 |
0.3445591595 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 17 |
Hb_001188_120 |
0.350104704 |
- |
- |
hypothetical protein JCGZ_05575 [Jatropha curcas] |
| 18 |
Hb_001206_010 |
0.3511121393 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 19 |
Hb_013080_010 |
0.354707648 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 20 |
Hb_005511_090 |
0.3576978628 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 C isoform X1 [Jatropha curcas] |