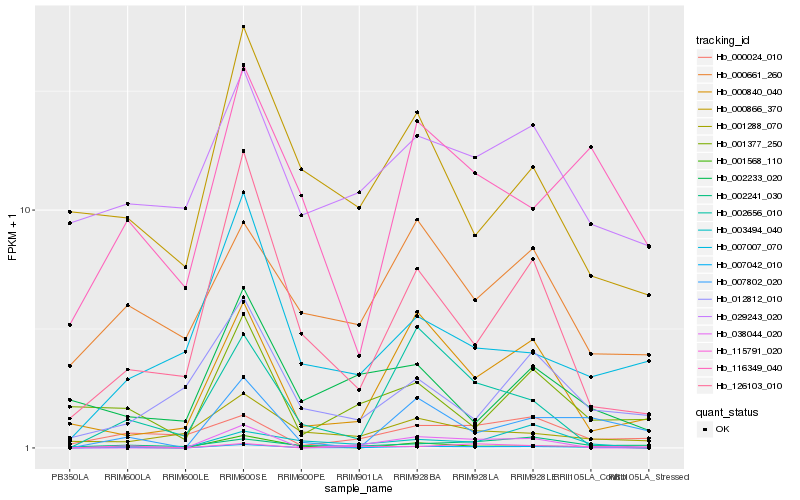
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007042_010 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 2 |
Hb_007802_020 |
0.2594228359 |
- |
- |
PREDICTED: receptor-like protein 12 [Eucalyptus grandis] |
| 3 |
Hb_003494_040 |
0.2633915465 |
transcription factor |
TF Family: NAC |
PREDICTED: protein FEZ [Jatropha curcas] |
| 4 |
Hb_126103_010 |
0.2656320158 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 5 |
Hb_001568_110 |
0.270218778 |
- |
- |
BnaA02g12660D [Brassica napus] |
| 6 |
Hb_116349_040 |
0.2774697032 |
transcription factor |
TF Family: Orphans |
PREDICTED: two-component response regulator ARR5 [Jatropha curcas] |
| 7 |
Hb_000840_040 |
0.2845330931 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 8 |
Hb_002656_010 |
0.2848839468 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_038044_020 |
0.2928330758 |
- |
- |
- |
| 10 |
Hb_115791_020 |
0.3092985788 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 11 |
Hb_002241_030 |
0.3103983556 |
- |
- |
Retrotransposon protein, putative [Theobroma cacao] |
| 12 |
Hb_007007_070 |
0.3107557992 |
transcription factor |
TF Family: TCP |
hypothetical protein MIMGU_mgv1a022855mg, partial [Erythranthe guttata] |
| 13 |
Hb_029243_020 |
0.3149091832 |
- |
- |
ENTH/VHS/GAT family protein [Theobroma cacao] |
| 14 |
Hb_001288_070 |
0.3165025963 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 15 |
Hb_001377_250 |
0.3176479466 |
- |
- |
dynamin, putative [Ricinus communis] |
| 16 |
Hb_000866_370 |
0.3182161894 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 17 |
Hb_000661_260 |
0.3191111802 |
- |
- |
PREDICTED: uncharacterized protein LOC105648304 [Jatropha curcas] |
| 18 |
Hb_012812_010 |
0.3234115174 |
- |
- |
putative leucine-rich repeat receptor-like serine/threonine-protein kinase [Morus notabilis] |
| 19 |
Hb_002233_020 |
0.3241850273 |
- |
- |
hypothetical protein JCGZ_04974 [Jatropha curcas] |
| 20 |
Hb_000024_010 |
0.3248299358 |
- |
- |
Uncharacterized protein TCM_044274 [Theobroma cacao] |