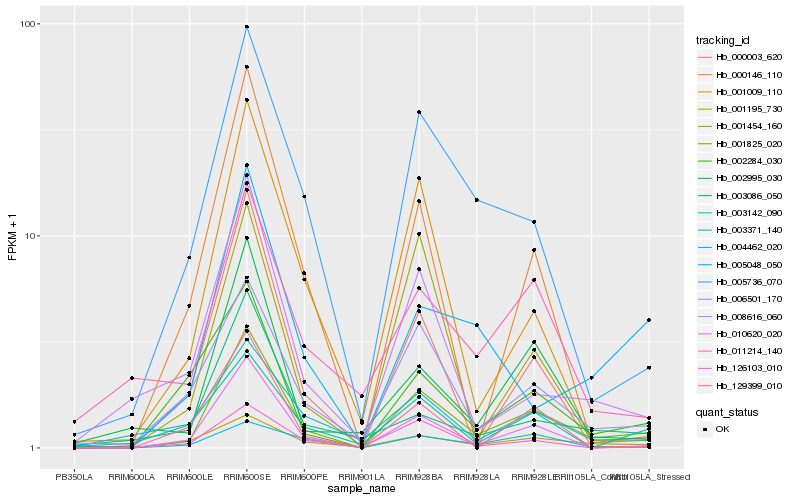
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001825_020 |
0.0 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 2 |
Hb_010620_020 |
0.1636884953 |
- |
- |
receptor protein kinase, putative [Ricinus communis] |
| 3 |
Hb_006501_170 |
0.1789378535 |
- |
- |
hypothetical protein JCGZ_07371 [Jatropha curcas] |
| 4 |
Hb_005736_070 |
0.1885431763 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 5 |
Hb_001195_730 |
0.2102609868 |
- |
- |
PREDICTED: protein LYK5-like [Jatropha curcas] |
| 6 |
Hb_002284_030 |
0.2103000319 |
- |
- |
PREDICTED: xylem cysteine proteinase 2 [Jatropha curcas] |
| 7 |
Hb_011214_140 |
0.215584949 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 8 |
Hb_004462_020 |
0.2168796045 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Vitis vinifera] |
| 9 |
Hb_002995_030 |
0.2177748784 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_003371_140 |
0.2211683397 |
- |
- |
hypothetical protein POPTR_0001s23540g, partial [Populus trichocarpa] |
| 11 |
Hb_008616_060 |
0.2214780689 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 12 |
Hb_003142_090 |
0.2217063352 |
- |
- |
Kinase, putative isoform 2 [Theobroma cacao] |
| 13 |
Hb_001009_110 |
0.2219373242 |
- |
- |
PREDICTED: uncharacterized protein LOC105640232 [Jatropha curcas] |
| 14 |
Hb_126103_010 |
0.2222587362 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 15 |
Hb_129399_010 |
0.2251938672 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_000146_110 |
0.2260902859 |
- |
- |
resveratrol/hydroxycinnamic acid O-glucosyltransferase [Vitis labrusca] |
| 17 |
Hb_003086_050 |
0.2274836062 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 18 |
Hb_000003_620 |
0.2282339049 |
- |
- |
PREDICTED: ABC transporter G family member 29-like [Jatropha curcas] |
| 19 |
Hb_005048_050 |
0.2295746105 |
- |
- |
Rotenone-insensitive NADH-ubiquinone oxidoreductase, mitochondrial precursor, putative [Ricinus communis] |
| 20 |
Hb_001454_160 |
0.2315423807 |
- |
- |
PREDICTED: translocase of chloroplast 159, chloroplastic [Jatropha curcas] |