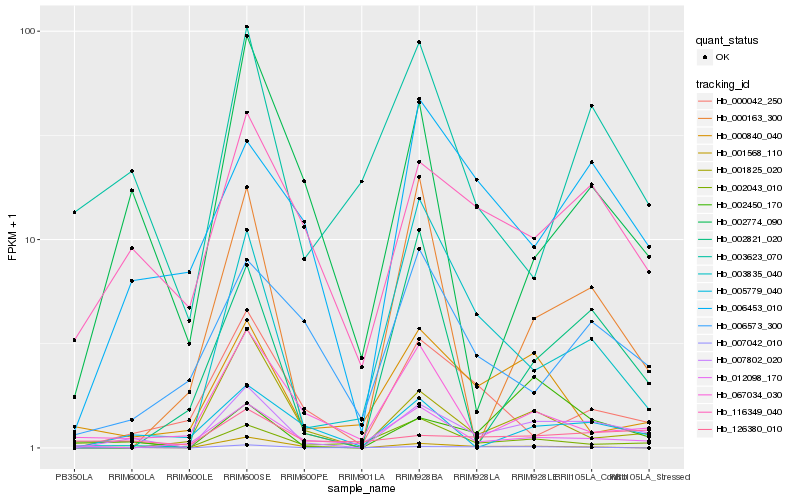
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007802_020 |
0.0 |
- |
- |
PREDICTED: receptor-like protein 12 [Eucalyptus grandis] |
| 2 |
Hb_116349_040 |
0.2417577091 |
transcription factor |
TF Family: Orphans |
PREDICTED: two-component response regulator ARR5 [Jatropha curcas] |
| 3 |
Hb_007042_010 |
0.2594228359 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 4 |
Hb_005779_040 |
0.2664429041 |
- |
- |
- |
| 5 |
Hb_006453_010 |
0.2727728189 |
- |
- |
PREDICTED: glutaredoxin-C9 [Jatropha curcas] |
| 6 |
Hb_012098_170 |
0.2809571735 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_16874 [Jatropha curcas] |
| 7 |
Hb_001825_020 |
0.2862694369 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 8 |
Hb_002774_090 |
0.2871710787 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_067034_030 |
0.2925929779 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 10 |
Hb_002043_010 |
0.2926381287 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 11 |
Hb_000042_250 |
0.2939491527 |
- |
- |
PREDICTED: uncharacterized protein LOC105632812 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001568_110 |
0.2980555266 |
- |
- |
BnaA02g12660D [Brassica napus] |
| 13 |
Hb_000840_040 |
0.2990468067 |
- |
- |
monoxygenase, putative [Ricinus communis] |
| 14 |
Hb_003623_070 |
0.3000379687 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_003835_040 |
0.3009957401 |
- |
- |
PREDICTED: UDP-glycosyltransferase 91A1-like [Jatropha curcas] |
| 16 |
Hb_006573_300 |
0.3014232521 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_126380_010 |
0.303522643 |
desease resistance |
Gene Name: NB-ARC |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_000163_300 |
0.3036949864 |
- |
- |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 19 |
Hb_002821_020 |
0.3048414078 |
desease resistance |
Gene Name: ABC_tran |
PREDICTED: ABC transporter C family member 4 [Vitis vinifera] |
| 20 |
Hb_002450_170 |
0.3061635142 |
transcription factor |
TF Family: FAR1 |
Protein FAR1-RELATED SEQUENCE 5 [Glycine soja] |