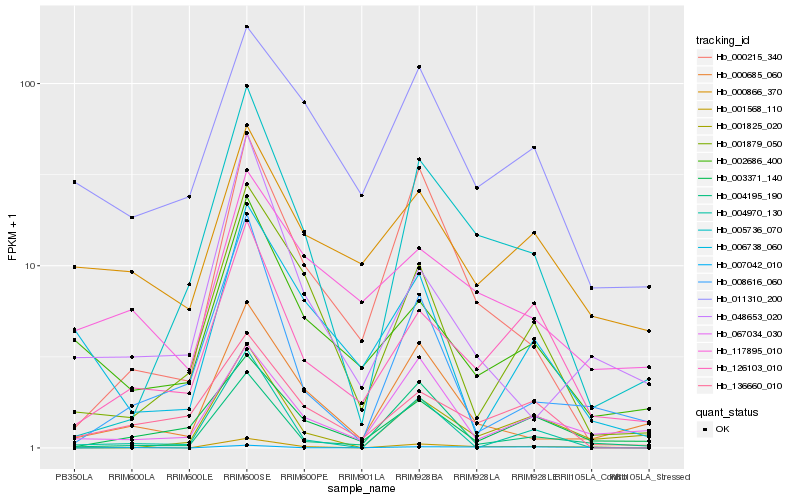
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001568_110 |
0.0 |
- |
- |
BnaA02g12660D [Brassica napus] |
| 2 |
Hb_126103_010 |
0.2415213968 |
- |
- |
Phosphoprotein phosphatase [Theobroma cacao] |
| 3 |
Hb_005736_070 |
0.248384058 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 4 |
Hb_067034_030 |
0.253174873 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 5 |
Hb_002686_400 |
0.2541208295 |
- |
- |
hypothetical protein POPTR_0019s04500g [Populus trichocarpa] |
| 6 |
Hb_001825_020 |
0.2584293919 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_000685_060 |
0.2601017789 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 8 |
Hb_000215_340 |
0.2667465228 |
- |
- |
gamma glutamyl transpeptidases, putative [Ricinus communis] |
| 9 |
Hb_007042_010 |
0.270218778 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 10 |
Hb_011310_200 |
0.2707306106 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 11 |
Hb_004195_190 |
0.2712707465 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 12 |
Hb_000866_370 |
0.2734885427 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 13 |
Hb_006738_060 |
0.2738320851 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 14 |
Hb_001879_050 |
0.2757100953 |
- |
- |
Disease resistance protein RFL1, putative [Ricinus communis] |
| 15 |
Hb_004970_130 |
0.2787615111 |
- |
- |
PREDICTED: uncharacterized protein LOC105633891 [Jatropha curcas] |
| 16 |
Hb_136660_010 |
0.2794858863 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 17 |
Hb_048653_020 |
0.2795594401 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_003371_140 |
0.2797173431 |
- |
- |
hypothetical protein POPTR_0001s23540g, partial [Populus trichocarpa] |
| 19 |
Hb_117895_010 |
0.2799266641 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |
| 20 |
Hb_008616_060 |
0.2804933472 |
- |
- |
syntaxin, putative [Ricinus communis] |