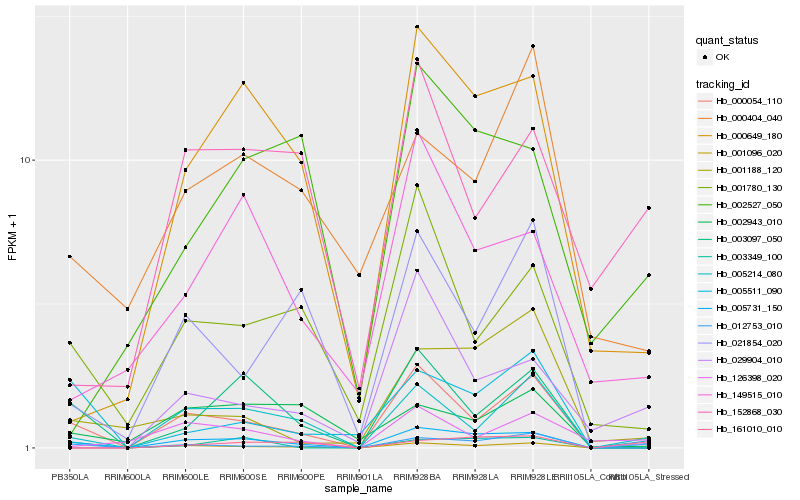
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005731_150 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105630360 [Jatropha curcas] |
| 2 |
Hb_000054_110 |
0.211777582 |
- |
- |
PREDICTED: uncharacterized protein LOC105350810 [Fragaria vesca subsp. vesca] |
| 3 |
Hb_005214_080 |
0.218429072 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_012753_010 |
0.237304732 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 5 |
Hb_000649_180 |
0.2465520487 |
- |
- |
PREDICTED: 1-aminocyclopropane-1-carboxylate oxidase homolog 4-like [Jatropha curcas] |
| 6 |
Hb_003097_050 |
0.2481015581 |
- |
- |
hypothetical protein POPTR_0007s07150g [Populus trichocarpa] |
| 7 |
Hb_001780_130 |
0.2511332777 |
- |
- |
hypothetical protein JCGZ_08545 [Jatropha curcas] |
| 8 |
Hb_005511_090 |
0.2633544826 |
- |
- |
PREDICTED: probable ubiquitin-conjugating enzyme E2 C isoform X1 [Jatropha curcas] |
| 9 |
Hb_149515_010 |
0.2682512347 |
- |
- |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 10 |
Hb_001096_020 |
0.2760880216 |
desease resistance |
Gene Name: TIR |
PREDICTED: TMV resistance protein N-like isoform X1 [Populus euphratica] |
| 11 |
Hb_003349_100 |
0.2783971251 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001188_120 |
0.2800888611 |
- |
- |
hypothetical protein JCGZ_05575 [Jatropha curcas] |
| 13 |
Hb_161010_010 |
0.285643291 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 14 |
Hb_152868_030 |
0.2880602948 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 15 |
Hb_021854_020 |
0.2906173888 |
- |
- |
hypothetical protein POPTR_1763s00200g, partial [Populus trichocarpa] |
| 16 |
Hb_029904_010 |
0.2928525149 |
- |
- |
PREDICTED: uncharacterized protein LOC105635075 [Jatropha curcas] |
| 17 |
Hb_000404_040 |
0.2929041222 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_126398_020 |
0.29346818 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_002527_050 |
0.2934852166 |
- |
- |
hypothetical protein JCGZ_20265 [Jatropha curcas] |
| 20 |
Hb_002943_010 |
0.2951496508 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Nelumbo nucifera] |