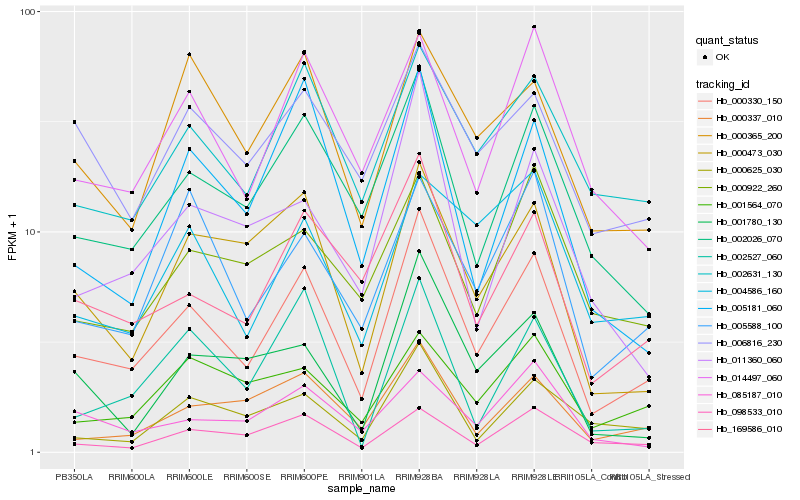
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000330_150 |
0.0 |
- |
- |
PREDICTED: dihydroflavonol-4-reductase-like [Jatropha curcas] |
| 2 |
Hb_002026_070 |
0.12419704 |
- |
- |
hypothetical protein PHAVU_002G2910001g, partial [Phaseolus vulgaris] |
| 3 |
Hb_169586_010 |
0.1253509194 |
- |
- |
Acid phosphatase/vanadium-dependent haloperoxidase-related protein [Theobroma cacao] |
| 4 |
Hb_002631_130 |
0.1383836127 |
- |
- |
PREDICTED: L-ascorbate oxidase-like [Jatropha curcas] |
| 5 |
Hb_014497_060 |
0.1408581235 |
- |
- |
phosphofructokinase, putative [Ricinus communis] |
| 6 |
Hb_000473_030 |
0.1425686265 |
- |
- |
hypothetical protein JCGZ_19297 [Jatropha curcas] |
| 7 |
Hb_000337_010 |
0.1450008086 |
- |
- |
PREDICTED: proteasome activator subunit 4 isoform X2 [Jatropha curcas] |
| 8 |
Hb_098533_010 |
0.1500742755 |
- |
- |
hypothetical protein L484_003492 [Morus notabilis] |
| 9 |
Hb_005181_060 |
0.1513392629 |
- |
- |
ATP-citrate synthase, putative [Ricinus communis] |
| 10 |
Hb_005588_100 |
0.153620671 |
- |
- |
protein phosphatase, putative [Ricinus communis] |
| 11 |
Hb_000365_200 |
0.1553691736 |
- |
- |
PREDICTED: 2,3-bisphosphoglycerate-independent phosphoglycerate mutase [Jatropha curcas] |
| 12 |
Hb_000625_030 |
0.1586668742 |
- |
- |
PREDICTED: calmodulin-lysine N-methyltransferase [Jatropha curcas] |
| 13 |
Hb_001564_070 |
0.1616910723 |
- |
- |
PREDICTED: G patch domain-containing protein TGH [Jatropha curcas] |
| 14 |
Hb_001780_130 |
0.1620462887 |
- |
- |
hypothetical protein JCGZ_08545 [Jatropha curcas] |
| 15 |
Hb_011360_060 |
0.1627730033 |
- |
- |
glycogen phosphorylase, putative [Ricinus communis] |
| 16 |
Hb_085187_010 |
0.1633963543 |
- |
- |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 17 |
Hb_000922_260 |
0.1638040525 |
- |
- |
PREDICTED: pantothenate kinase 2 [Jatropha curcas] |
| 18 |
Hb_004586_160 |
0.1643056827 |
- |
- |
PREDICTED: glucose-6-phosphate isomerase 1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_006816_230 |
0.1645960089 |
- |
- |
protein disulfide isomerase, putative [Ricinus communis] |
| 20 |
Hb_002527_060 |
0.1656547273 |
- |
- |
PREDICTED: protein NRT1/ PTR FAMILY 5.10-like [Jatropha curcas] |