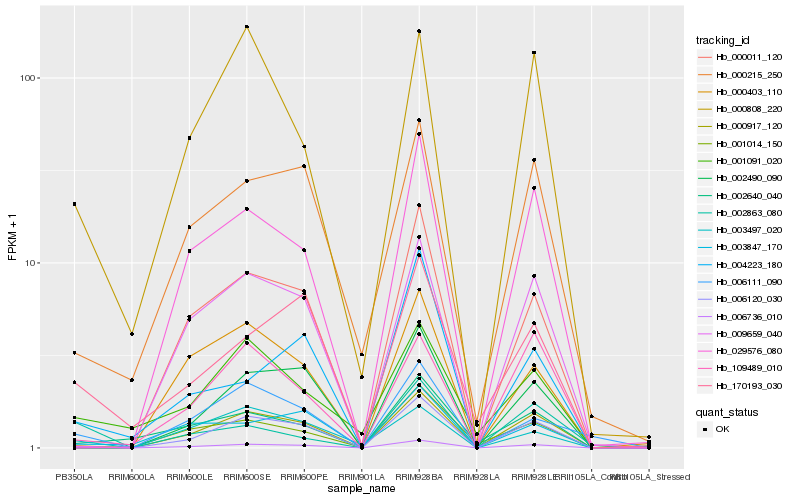
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006736_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_170193_030 |
0.1830172138 |
- |
- |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 3 |
Hb_006111_090 |
0.1843674786 |
- |
- |
hypothetical protein JCGZ_26825 [Jatropha curcas] |
| 4 |
Hb_002863_080 |
0.1946894014 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: probable disease resistance protein At5g63020 [Jatropha curcas] |
| 5 |
Hb_000808_220 |
0.2006083025 |
- |
- |
PREDICTED: pyrophosphate-energized vacuolar membrane proton pump 1-like [Jatropha curcas] |
| 6 |
Hb_003497_020 |
0.2015653444 |
transcription factor |
TF Family: MYB |
MYB transcriptional factor [Populus nigra] |
| 7 |
Hb_000215_250 |
0.2076558884 |
- |
- |
Acid phosphatase 1 precursor, putative [Ricinus communis] |
| 8 |
Hb_001091_020 |
0.211012313 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RGA2-like [Jatropha curcas] |
| 9 |
Hb_000917_120 |
0.2117632725 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_006120_030 |
0.2128717346 |
- |
- |
- |
| 11 |
Hb_003847_170 |
0.2166233032 |
- |
- |
PREDICTED: trehalase isoform X1 [Jatropha curcas] |
| 12 |
Hb_000011_120 |
0.2168480114 |
- |
- |
PREDICTED: ATPase 10, plasma membrane-type-like [Populus euphratica] |
| 13 |
Hb_009659_040 |
0.2203243131 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER [Populus euphratica] |
| 14 |
Hb_002640_040 |
0.222418914 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_029576_080 |
0.2245770576 |
- |
- |
Aspartic proteinase precursor, putative [Ricinus communis] |
| 16 |
Hb_001014_150 |
0.2259432488 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 17 |
Hb_002490_090 |
0.2270422861 |
- |
- |
PREDICTED: uncharacterized protein LOC105133003 [Populus euphratica] |
| 18 |
Hb_109489_010 |
0.2302610736 |
- |
- |
multidrug resistance-associated protein 2, 6 (mrp2, 6), abc-transoprter, putative [Ricinus communis] |
| 19 |
Hb_000403_110 |
0.23044053 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 51 [Jatropha curcas] |
| 20 |
Hb_004223_180 |
0.2329426541 |
- |
- |
PREDICTED: auxin-induced protein 15A [Jatropha curcas] |