| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
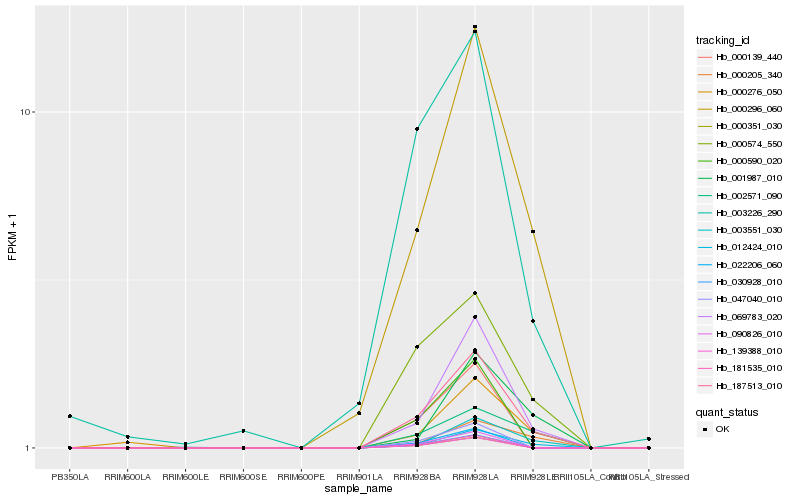
Hb_187513_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105761922 [Gossypium raimondii] |
| 2 |
Hb_069783_020 |
0.1008270522 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647845 [Jatropha curcas] |
| 3 |
Hb_000296_060 |
0.108156474 |
- |
- |
- |
| 4 |
Hb_000574_550 |
0.1382471846 |
- |
- |
PREDICTED: SNF1-related protein kinase regulatory subunit gamma-like PV42a [Jatropha curcas] |
| 5 |
Hb_002571_090 |
0.1589706895 |
- |
- |
- |
| 6 |
Hb_012424_010 |
0.1591202101 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 7 |
Hb_003551_030 |
0.17479796 |
- |
- |
hypothetical protein POPTR_0016s03850g [Populus trichocarpa] |
| 8 |
Hb_000276_050 |
0.1820479742 |
- |
- |
- |
| 9 |
Hb_003226_290 |
0.1837271873 |
- |
- |
catalytic, putative [Ricinus communis] |
| 10 |
Hb_000205_340 |
0.1951163553 |
- |
- |
PREDICTED: putative B3 domain-containing protein At2g27410 [Jatropha curcas] |
| 11 |
Hb_047040_010 |
0.2152744825 |
- |
- |
integrase [Gossypium herbaceum] |
| 12 |
Hb_000590_020 |
0.215275922 |
- |
- |
hypothetical protein JCGZ_18748 [Jatropha curcas] |
| 13 |
Hb_022206_060 |
0.2155095728 |
- |
- |
- |
| 14 |
Hb_000139_440 |
0.2156496206 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_090826_010 |
0.2157562237 |
- |
- |
PREDICTED: uncharacterized protein LOC105786910 [Gossypium raimondii] |
| 16 |
Hb_001987_010 |
0.2159760877 |
- |
- |
- |
| 17 |
Hb_030928_010 |
0.2161178184 |
- |
- |
- |
| 18 |
Hb_181535_010 |
0.2161794035 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 19 |
Hb_000351_030 |
0.2164050042 |
- |
- |
putative NBS-LRR type disease resistance protein [Prunus persica] |
| 20 |
Hb_139388_010 |
0.216436929 |
- |
- |
PREDICTED: uncharacterized protein LOC104222982, partial [Nicotiana sylvestris] |