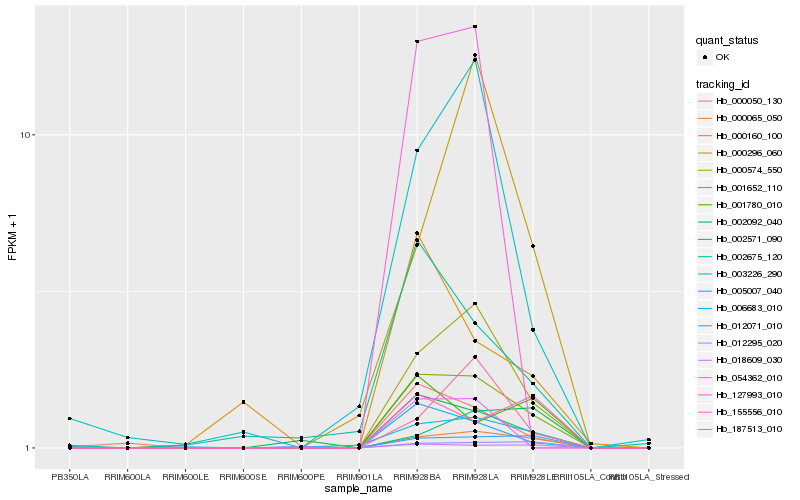
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001652_110 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104248159 [Nicotiana sylvestris] |
| 2 |
Hb_000065_050 |
0.1280657089 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 3 |
Hb_000574_550 |
0.1388722583 |
- |
- |
PREDICTED: SNF1-related protein kinase regulatory subunit gamma-like PV42a [Jatropha curcas] |
| 4 |
Hb_005007_040 |
0.1994637016 |
- |
- |
hypothetical protein VITISV_032357 [Vitis vinifera] |
| 5 |
Hb_012071_010 |
0.2043644978 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 6 |
Hb_006683_010 |
0.2076743816 |
- |
- |
cell wall invertase [Manihot esculenta] |
| 7 |
Hb_012295_020 |
0.2103486697 |
- |
- |
PREDICTED: uncharacterized protein LOC104223093 [Nicotiana sylvestris] |
| 8 |
Hb_000050_130 |
0.2115203652 |
- |
- |
Putative Pol polyprotein from transposon element Bs1 [Triticum urartu] |
| 9 |
Hb_002092_040 |
0.222767445 |
- |
- |
PREDICTED: protein YLS3 [Jatropha curcas] |
| 10 |
Hb_002571_090 |
0.229013094 |
- |
- |
- |
| 11 |
Hb_002675_120 |
0.246904417 |
- |
- |
DNA-repair protein xrcc1, putative [Ricinus communis] |
| 12 |
Hb_003226_290 |
0.2470530015 |
- |
- |
catalytic, putative [Ricinus communis] |
| 13 |
Hb_155556_010 |
0.2659024012 |
- |
- |
putative methylenetetrahydrofolate reductase [Morus notabilis] |
| 14 |
Hb_001780_010 |
0.2660352428 |
- |
- |
hypothetical protein CICLE_v10006597mg [Citrus clementina] |
| 15 |
Hb_187513_010 |
0.2745314091 |
- |
- |
PREDICTED: uncharacterized protein LOC105761922 [Gossypium raimondii] |
| 16 |
Hb_018609_030 |
0.2862683375 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000160_100 |
0.2887900902 |
- |
- |
PREDICTED: uncharacterized transporter YBR287W-like [Populus euphratica] |
| 18 |
Hb_054362_010 |
0.293748861 |
- |
- |
PREDICTED: uncharacterized protein LOC105650428 [Jatropha curcas] |
| 19 |
Hb_000296_060 |
0.2944345544 |
- |
- |
- |
| 20 |
Hb_127993_010 |
0.2951296978 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X4 [Jatropha curcas] |