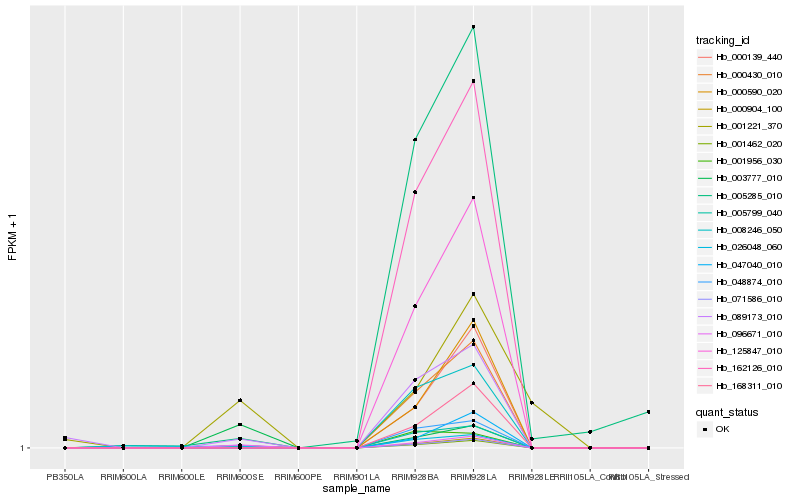
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000904_100 |
0.0 |
- |
- |
hypothetical protein JCGZ_03883 [Jatropha curcas] |
| 2 |
Hb_096671_010 |
0.0021310726 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 3 |
Hb_001462_020 |
0.0277729754 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 4 |
Hb_071586_010 |
0.0281926587 |
- |
- |
PREDICTED: uncharacterized protein LOC105632410 [Jatropha curcas] |
| 5 |
Hb_001956_030 |
0.2269317249 |
- |
- |
PREDICTED: xyloglucan galactosyltransferase KATAMARI1 homolog [Populus euphratica] |
| 6 |
Hb_089173_010 |
0.2285147269 |
- |
- |
PREDICTED: uncharacterized protein LOC105636883 [Jatropha curcas] |
| 7 |
Hb_003777_010 |
0.2585329479 |
- |
- |
- |
| 8 |
Hb_125847_010 |
0.2790496623 |
- |
- |
- |
| 9 |
Hb_000430_010 |
0.2793185229 |
- |
- |
- |
| 10 |
Hb_162126_010 |
0.2807646693 |
- |
- |
- |
| 11 |
Hb_001221_370 |
0.2843969508 |
- |
- |
hypothetical protein MIMGU_mgv1a020064mg, partial [Erythranthe guttata] |
| 12 |
Hb_168311_010 |
0.2849771401 |
- |
- |
- |
| 13 |
Hb_005285_010 |
0.2876963037 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 14 |
Hb_000139_440 |
0.2901969961 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_026048_060 |
0.2902870517 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 16 |
Hb_000590_020 |
0.2932168546 |
- |
- |
hypothetical protein JCGZ_18748 [Jatropha curcas] |
| 17 |
Hb_008246_050 |
0.2936745009 |
- |
- |
- |
| 18 |
Hb_005799_040 |
0.2936882531 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET4-like isoform X1 [Gossypium raimondii] |
| 19 |
Hb_047040_010 |
0.2946134216 |
- |
- |
integrase [Gossypium herbaceum] |
| 20 |
Hb_048874_010 |
0.2956125085 |
- |
- |
hypothetical protein POPTR_0016s10870g [Populus trichocarpa] |