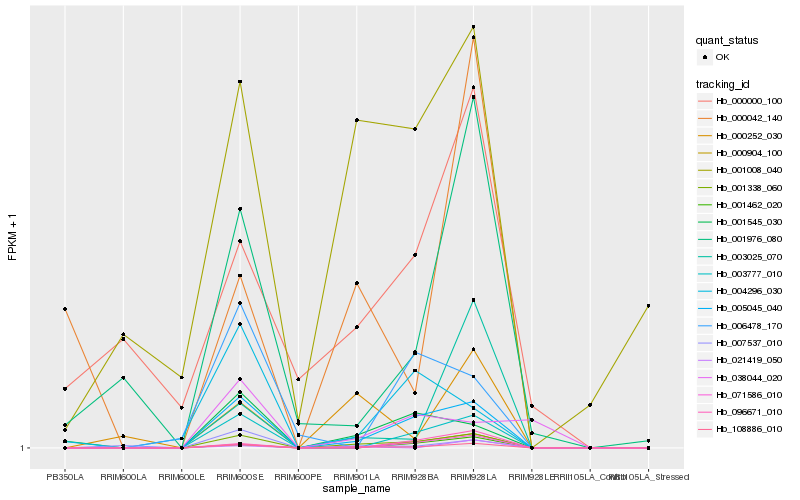
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001338_060 |
0.0 |
- |
- |
- |
| 2 |
Hb_108886_010 |
0.0984114493 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_005045_040 |
0.1137964279 |
- |
- |
hypothetical protein POPTR_0017s04090g, partial [Populus trichocarpa] |
| 4 |
Hb_003777_010 |
0.2312182991 |
- |
- |
- |
| 5 |
Hb_001545_030 |
0.2592712569 |
- |
- |
- |
| 6 |
Hb_071586_010 |
0.3031376442 |
- |
- |
PREDICTED: uncharacterized protein LOC105632410 [Jatropha curcas] |
| 7 |
Hb_001462_020 |
0.3034352047 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 8 |
Hb_007537_010 |
0.3066941429 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 9 |
Hb_000252_030 |
0.3087952981 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 10 |
Hb_001976_080 |
0.3144037243 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor AIL6 [Jatropha curcas] |
| 11 |
Hb_003025_070 |
0.3171368846 |
- |
- |
PREDICTED: uncharacterized protein LOC103419743 [Malus domestica] |
| 12 |
Hb_001008_040 |
0.3171772582 |
- |
- |
Late embryogenesis abundant protein D-7, putative [Ricinus communis] |
| 13 |
Hb_096671_010 |
0.3207381601 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 14 |
Hb_004296_030 |
0.3211324877 |
- |
- |
PREDICTED: gamma-tubulin complex component 3 [Jatropha curcas] |
| 15 |
Hb_000904_100 |
0.3212750122 |
- |
- |
hypothetical protein JCGZ_03883 [Jatropha curcas] |
| 16 |
Hb_006478_170 |
0.3435557444 |
- |
- |
hypothetical protein POPTR_0005s23360g [Populus trichocarpa] |
| 17 |
Hb_000000_100 |
0.3547373349 |
- |
- |
hypothetical protein POPTR_0002s13260g [Populus trichocarpa] |
| 18 |
Hb_000042_140 |
0.3595419582 |
- |
- |
hypothetical protein POPTR_0001s41640g, partial [Populus trichocarpa] |
| 19 |
Hb_021419_050 |
0.360220952 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Malus domestica] |
| 20 |
Hb_038044_020 |
0.3645268739 |
- |
- |
- |