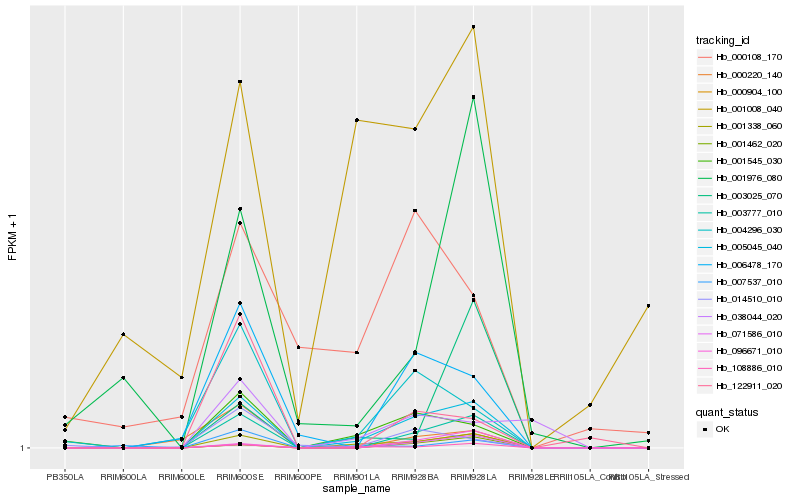
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005045_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0017s04090g, partial [Populus trichocarpa] |
| 2 |
Hb_001338_060 |
0.1137964279 |
- |
- |
- |
| 3 |
Hb_003777_010 |
0.1636790691 |
- |
- |
- |
| 4 |
Hb_108886_010 |
0.1982300326 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_001545_030 |
0.2300105935 |
- |
- |
- |
| 6 |
Hb_004296_030 |
0.2575076306 |
- |
- |
PREDICTED: gamma-tubulin complex component 3 [Jatropha curcas] |
| 7 |
Hb_006478_170 |
0.2641603779 |
- |
- |
hypothetical protein POPTR_0005s23360g [Populus trichocarpa] |
| 8 |
Hb_071586_010 |
0.2749347675 |
- |
- |
PREDICTED: uncharacterized protein LOC105632410 [Jatropha curcas] |
| 9 |
Hb_001462_020 |
0.2755308687 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 10 |
Hb_096671_010 |
0.2954494581 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 11 |
Hb_000904_100 |
0.2965259678 |
- |
- |
hypothetical protein JCGZ_03883 [Jatropha curcas] |
| 12 |
Hb_007537_010 |
0.3155546742 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 13 |
Hb_014510_010 |
0.3162965409 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |
| 14 |
Hb_000220_140 |
0.322349675 |
- |
- |
palmitoyl-acyl carrier protein thioesterase [Ricinus communis] |
| 15 |
Hb_001976_080 |
0.3224621805 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor AIL6 [Jatropha curcas] |
| 16 |
Hb_038044_020 |
0.3365024141 |
- |
- |
- |
| 17 |
Hb_122911_020 |
0.342058712 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 18 |
Hb_001008_040 |
0.3578730719 |
- |
- |
Late embryogenesis abundant protein D-7, putative [Ricinus communis] |
| 19 |
Hb_003025_070 |
0.368598134 |
- |
- |
PREDICTED: uncharacterized protein LOC103419743 [Malus domestica] |
| 20 |
Hb_000108_170 |
0.3707768018 |
- |
- |
PREDICTED: cytokinin riboside 5'-monophosphate phosphoribohydrolase LOG1-like [Jatropha curcas] |