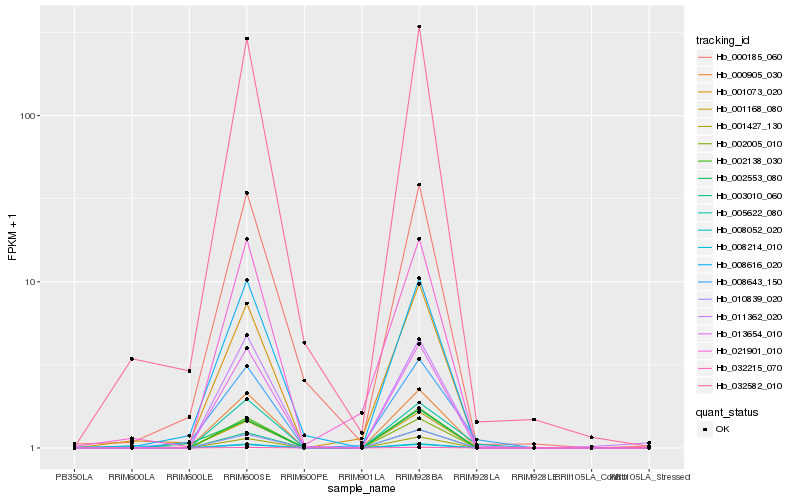
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008643_150 |
0.0 |
- |
- |
hypothetical protein CISIN_1g0034652mg, partial [Citrus sinensis] |
| 2 |
Hb_000905_030 |
0.1403327038 |
- |
- |
PREDICTED: cytochrome P450 87A3-like [Jatropha curcas] |
| 3 |
Hb_001168_080 |
0.1416071739 |
- |
- |
kinase, putative [Ricinus communis] |
| 4 |
Hb_001073_020 |
0.1424473993 |
- |
- |
hypothetical protein CISIN_1g031854mg [Citrus sinensis] |
| 5 |
Hb_005622_080 |
0.1500342915 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 6 |
Hb_032582_010 |
0.1500764485 |
- |
- |
PREDICTED: cytochrome P450 CYP82D47-like [Jatropha curcas] |
| 7 |
Hb_008616_020 |
0.1523925902 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 8 |
Hb_032215_070 |
0.1584073647 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 9 |
Hb_001427_130 |
0.1584845936 |
- |
- |
PREDICTED: uncharacterized protein LOC101510272 [Cicer arietinum] |
| 10 |
Hb_008052_020 |
0.1588720093 |
- |
- |
PREDICTED: proline-rich receptor-like protein kinase PERK15 [Brassica rapa] |
| 11 |
Hb_008214_010 |
0.1589129245 |
- |
- |
PREDICTED: uncharacterized protein LOC105797726 [Gossypium raimondii] |
| 12 |
Hb_003010_060 |
0.1590222074 |
- |
- |
Patatin T5 precursor, putative [Ricinus communis] |
| 13 |
Hb_002553_080 |
0.1592284905 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF096-like [Eucalyptus grandis] |
| 14 |
Hb_002005_010 |
0.159377493 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB39-like isoform X2 [Populus euphratica] |
| 15 |
Hb_021901_010 |
0.1602271251 |
- |
- |
- |
| 16 |
Hb_000185_060 |
0.1612077897 |
- |
- |
PREDICTED: short-chain dehydrogenase TIC 32, chloroplastic [Jatropha curcas] |
| 17 |
Hb_013654_010 |
0.1618827596 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_011362_020 |
0.1619785237 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_010839_020 |
0.1632543515 |
- |
- |
- |
| 20 |
Hb_002138_030 |
0.1638304167 |
- |
- |
- |