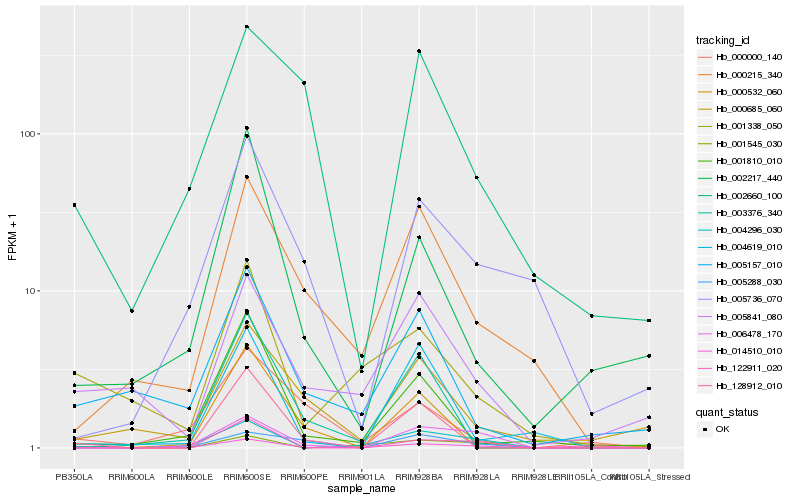
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_014510_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |
| 2 |
Hb_004296_030 |
0.1636672685 |
- |
- |
PREDICTED: gamma-tubulin complex component 3 [Jatropha curcas] |
| 3 |
Hb_006478_170 |
0.1933675497 |
- |
- |
hypothetical protein POPTR_0005s23360g [Populus trichocarpa] |
| 4 |
Hb_000215_340 |
0.2316967288 |
- |
- |
gamma glutamyl transpeptidases, putative [Ricinus communis] |
| 5 |
Hb_001545_030 |
0.2349686639 |
- |
- |
- |
| 6 |
Hb_005841_080 |
0.2481913713 |
- |
- |
hypothetical protein POPTR_0018s11370g [Populus trichocarpa] |
| 7 |
Hb_001338_050 |
0.2609148186 |
- |
- |
hypothetical protein POPTR_0016s05400g [Populus trichocarpa] |
| 8 |
Hb_005736_070 |
0.2643259455 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 9 |
Hb_001810_010 |
0.2666836097 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Nelumbo nucifera] |
| 10 |
Hb_005157_010 |
0.2700161277 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 11 |
Hb_005288_030 |
0.2766226175 |
- |
- |
PREDICTED: uncharacterized protein LOC105647760 [Jatropha curcas] |
| 12 |
Hb_128912_010 |
0.2770182142 |
- |
- |
PREDICTED: probable disease resistance protein At4g27220 [Jatropha curcas] |
| 13 |
Hb_000685_060 |
0.2776652502 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 14 |
Hb_002660_100 |
0.2821510851 |
- |
- |
plasma membrane intrinsic protein [Hevea brasiliensis] |
| 15 |
Hb_122911_020 |
0.284033411 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 16 |
Hb_003376_340 |
0.284524243 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002217_440 |
0.2864408299 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 18 |
Hb_004619_010 |
0.2876626557 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 19 |
Hb_000000_140 |
0.2890158841 |
- |
- |
- |
| 20 |
Hb_000532_060 |
0.2908786013 |
- |
- |
copper-transporting atpase p-type, putative [Ricinus communis] |