| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
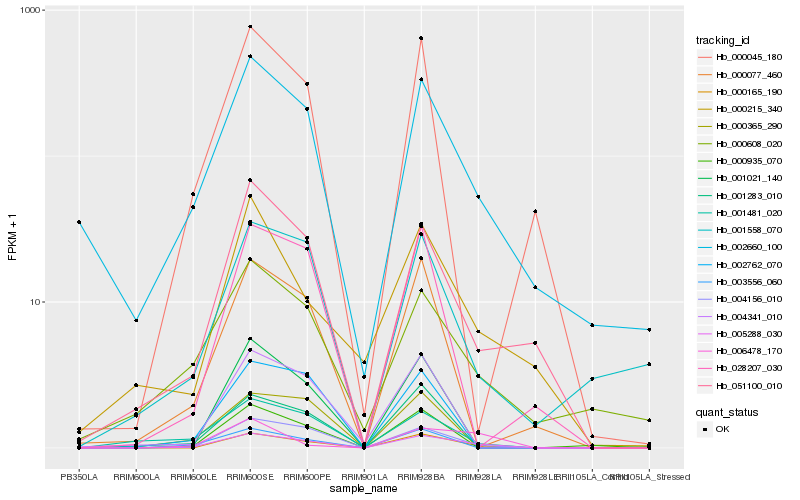
Hb_005288_030 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105647760 [Jatropha curcas] |
| 2 |
Hb_004156_010 |
0.1588635801 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |
| 3 |
Hb_000165_190 |
0.173251095 |
- |
- |
PREDICTED: U-box domain-containing protein 12-like [Jatropha curcas] |
| 4 |
Hb_002660_100 |
0.1929095963 |
- |
- |
plasma membrane intrinsic protein [Hevea brasiliensis] |
| 5 |
Hb_051100_010 |
0.2063794756 |
- |
- |
PREDICTED: putative UDP-rhamnose:rhamnosyltransferase 1 [Jatropha curcas] |
| 6 |
Hb_001481_020 |
0.2158543965 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase V.9-like [Jatropha curcas] |
| 7 |
Hb_000608_020 |
0.2252147866 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Jatropha curcas] |
| 8 |
Hb_001558_070 |
0.2322755224 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 9 |
Hb_003556_060 |
0.2331725499 |
- |
- |
- |
| 10 |
Hb_000077_460 |
0.2355451826 |
- |
- |
- |
| 11 |
Hb_006478_170 |
0.236704822 |
- |
- |
hypothetical protein POPTR_0005s23360g [Populus trichocarpa] |
| 12 |
Hb_000215_340 |
0.2369718325 |
- |
- |
gamma glutamyl transpeptidases, putative [Ricinus communis] |
| 13 |
Hb_001021_140 |
0.2398175062 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 14 |
Hb_000935_070 |
0.2410566142 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 15 |
Hb_002762_070 |
0.2416433252 |
transcription factor |
TF Family: HB |
homeobox protein knotted-1, putative [Ricinus communis] |
| 16 |
Hb_001283_010 |
0.2425264834 |
transcription factor |
TF Family: LOB |
LOB domain-containing protein 15 [Populus tremula x Populus tremuloides] |
| 17 |
Hb_000045_180 |
0.2433430053 |
- |
- |
aquaporin PIP1 [Hevea brasiliensis] |
| 18 |
Hb_004341_010 |
0.2447962818 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 19 |
Hb_028207_030 |
0.2450289431 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 8-like [Citrus sinensis] |
| 20 |
Hb_000365_290 |
0.2469805373 |
- |
- |
PREDICTED: protein MTL1-like isoform X1 [Jatropha curcas] |