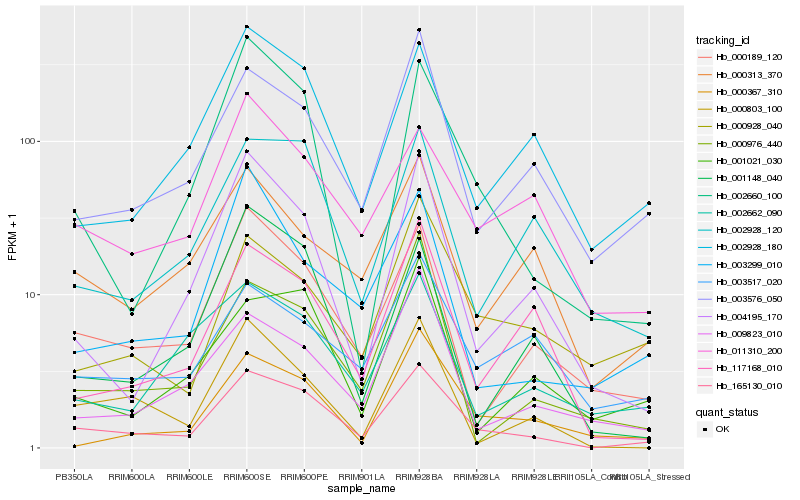
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_165130_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105648304 [Jatropha curcas] |
| 2 |
Hb_000189_120 |
0.1663361458 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE12-like isoform X5 [Jatropha curcas] |
| 3 |
Hb_003576_050 |
0.1696859433 |
transcription factor |
TF Family: EIL |
ETHYLENE-INSENSITIVE3 protein, putative [Ricinus communis] |
| 4 |
Hb_002660_100 |
0.1763189411 |
- |
- |
plasma membrane intrinsic protein [Hevea brasiliensis] |
| 5 |
Hb_002928_180 |
0.1775813552 |
- |
- |
catalase CAT1 [Manihot esculenta] |
| 6 |
Hb_002928_120 |
0.1820625172 |
- |
- |
1,4-alpha-glucan branching enzyme [Manihot esculenta] |
| 7 |
Hb_011310_200 |
0.1825487813 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 8 |
Hb_000803_100 |
0.1856462795 |
- |
- |
PREDICTED: uncharacterized protein LOC105648304 [Jatropha curcas] |
| 9 |
Hb_004195_170 |
0.1885276173 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_117168_010 |
0.1887423104 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 [Jatropha curcas] |
| 11 |
Hb_000313_370 |
0.190086227 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_003517_020 |
0.1949194197 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: ABC transporter C family member 3-like [Jatropha curcas] |
| 13 |
Hb_000367_310 |
0.1949244343 |
- |
- |
Brassinosteroid LRR receptor kinase precursor, putative [Ricinus communis] |
| 14 |
Hb_000928_040 |
0.1978707765 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_001021_030 |
0.1981083581 |
- |
- |
chloroplast-targeted copper chaperone, putative [Ricinus communis] |
| 16 |
Hb_002662_090 |
0.200635523 |
- |
- |
PREDICTED: probable inactive receptor kinase At5g67200 [Jatropha curcas] |
| 17 |
Hb_001148_040 |
0.2014006319 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 18 |
Hb_000976_440 |
0.2017258766 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 19 |
Hb_009823_010 |
0.2029802796 |
- |
- |
PREDICTED: uncharacterized protein LOC105633096 [Jatropha curcas] |
| 20 |
Hb_003299_010 |
0.2057487252 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |