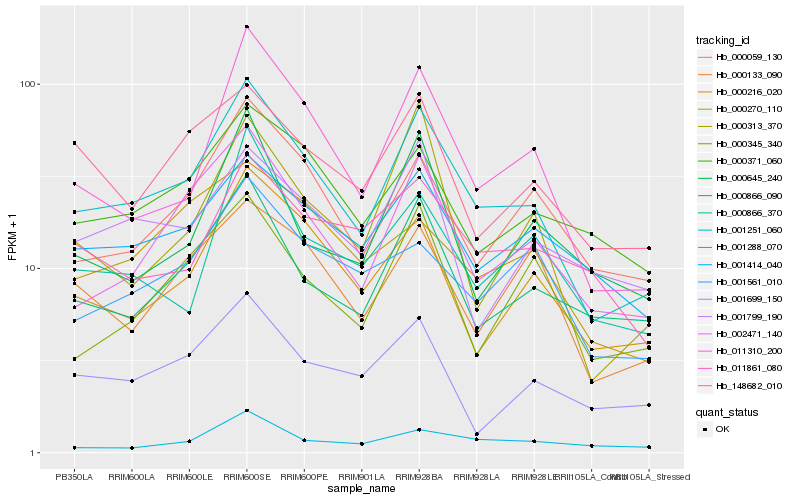
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001251_060 |
0.0 |
- |
- |
aldo/keto reductase AKR [Manihot esculenta] |
| 2 |
Hb_002471_140 |
0.1139848575 |
- |
- |
PREDICTED: protein OBERON 3 [Jatropha curcas] |
| 3 |
Hb_011310_200 |
0.1170048758 |
- |
- |
grr1, plant, putative [Ricinus communis] |
| 4 |
Hb_000216_020 |
0.1280843479 |
- |
- |
PREDICTED: benzaldehyde dehydrogenase (NAD(+))-like [Jatropha curcas] |
| 5 |
Hb_000866_090 |
0.1296253558 |
- |
- |
PREDICTED: U-box domain-containing protein 33-like isoform X3 [Jatropha curcas] |
| 6 |
Hb_000059_130 |
0.13062119 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 7 |
Hb_001414_040 |
0.1349992955 |
- |
- |
PREDICTED: RNA-dependent RNA polymerase 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000371_060 |
0.1376512582 |
transcription factor |
TF Family: BES1 |
BRASSINAZOLE-RESISTANT 1 protein, putative [Ricinus communis] |
| 9 |
Hb_148682_010 |
0.1466022752 |
- |
- |
PREDICTED: uncharacterized protein LOC105630720 [Jatropha curcas] |
| 10 |
Hb_000133_090 |
0.1467837015 |
- |
- |
PREDICTED: uncharacterized protein LOC105629320 [Jatropha curcas] |
| 11 |
Hb_000866_370 |
0.1503222452 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 12 |
Hb_001561_010 |
0.150787571 |
- |
- |
PREDICTED: uncharacterized protein LOC105635045 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000345_340 |
0.1513004001 |
- |
- |
PREDICTED: DNA polymerase zeta catalytic subunit [Jatropha curcas] |
| 14 |
Hb_000270_110 |
0.153106537 |
- |
- |
PREDICTED: calcium-dependent protein kinase 10 [Jatropha curcas] |
| 15 |
Hb_001699_150 |
0.1543471594 |
- |
- |
hypothetical protein POPTR_0009s13450g [Populus trichocarpa] |
| 16 |
Hb_001288_070 |
0.1548797087 |
- |
- |
PREDICTED: aminopeptidase M1-like [Jatropha curcas] |
| 17 |
Hb_001799_190 |
0.1555276219 |
- |
- |
PREDICTED: uncharacterized protein LOC105643221 [Jatropha curcas] |
| 18 |
Hb_000313_370 |
0.1568791525 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 19 |
Hb_011861_080 |
0.1568939727 |
- |
- |
hypothetical protein JCGZ_06088 [Jatropha curcas] |
| 20 |
Hb_000645_240 |
0.1571209606 |
- |
- |
AMP dependent ligase, putative [Ricinus communis] |