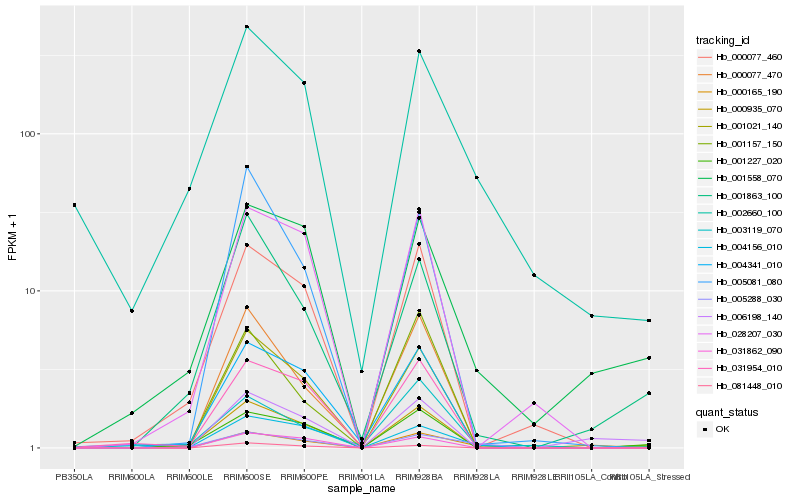
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000165_190 |
0.0 |
- |
- |
PREDICTED: U-box domain-containing protein 12-like [Jatropha curcas] |
| 2 |
Hb_005288_030 |
0.173251095 |
- |
- |
PREDICTED: uncharacterized protein LOC105647760 [Jatropha curcas] |
| 3 |
Hb_004156_010 |
0.1821243604 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |
| 4 |
Hb_001021_140 |
0.1932008592 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein indeterminate-domain 2-like [Jatropha curcas] |
| 5 |
Hb_001227_020 |
0.1975490922 |
- |
- |
- |
| 6 |
Hb_001558_070 |
0.2068640878 |
- |
- |
PREDICTED: mitochondrial uncoupling protein 5 [Jatropha curcas] |
| 7 |
Hb_000077_470 |
0.214116228 |
- |
- |
PREDICTED: anthranilate N-methyltransferase-like [Jatropha curcas] |
| 8 |
Hb_001863_100 |
0.218225812 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_031862_090 |
0.2209221301 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 10 |
Hb_006198_140 |
0.2214739109 |
- |
- |
hypothetical protein L484_015208 [Morus notabilis] |
| 11 |
Hb_031954_010 |
0.2243235785 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_004341_010 |
0.2259945397 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g61370 [Jatropha curcas] |
| 13 |
Hb_003119_070 |
0.2265012465 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 14 |
Hb_081448_010 |
0.2274997389 |
- |
- |
hypothetical protein JCGZ_11284 [Jatropha curcas] |
| 15 |
Hb_001157_150 |
0.2283477604 |
- |
- |
PREDICTED: transmembrane protein 45A-like [Jatropha curcas] |
| 16 |
Hb_000935_070 |
0.2288098245 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 17 |
Hb_005081_080 |
0.2308639917 |
- |
- |
hypothetical protein JCGZ_16787 [Jatropha curcas] |
| 18 |
Hb_028207_030 |
0.2320171678 |
- |
- |
PREDICTED: non-specific lipid-transfer protein 8-like [Citrus sinensis] |
| 19 |
Hb_000077_460 |
0.2329743285 |
- |
- |
- |
| 20 |
Hb_002660_100 |
0.2332982593 |
- |
- |
plasma membrane intrinsic protein [Hevea brasiliensis] |