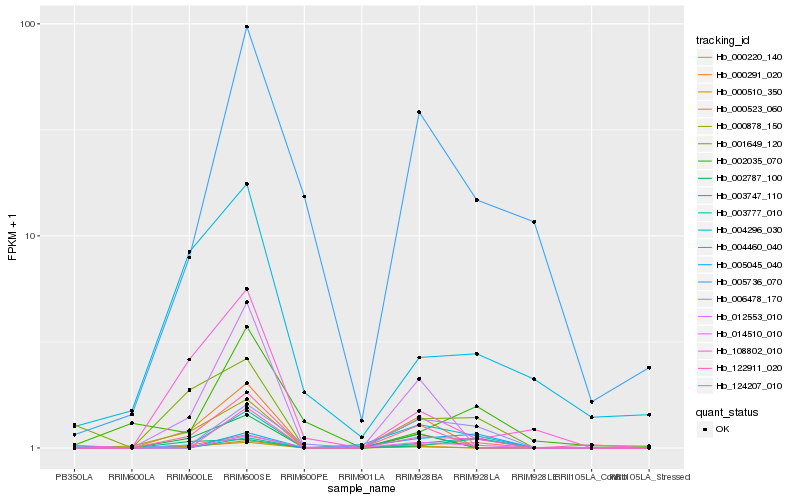
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000220_140 |
0.0 |
- |
- |
palmitoyl-acyl carrier protein thioesterase [Ricinus communis] |
| 2 |
Hb_006478_170 |
0.236752726 |
- |
- |
hypothetical protein POPTR_0005s23360g [Populus trichocarpa] |
| 3 |
Hb_003747_110 |
0.2495481978 |
- |
- |
PREDICTED: xyloglucan endotransglucosylase/hydrolase 1 [Jatropha curcas] |
| 4 |
Hb_004296_030 |
0.2612674794 |
- |
- |
PREDICTED: gamma-tubulin complex component 3 [Jatropha curcas] |
| 5 |
Hb_001649_120 |
0.2816716925 |
- |
- |
- |
| 6 |
Hb_003777_010 |
0.2843830259 |
- |
- |
- |
| 7 |
Hb_122911_020 |
0.289504822 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 8 |
Hb_014510_010 |
0.3008044918 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |
| 9 |
Hb_005045_040 |
0.322349675 |
- |
- |
hypothetical protein POPTR_0017s04090g, partial [Populus trichocarpa] |
| 10 |
Hb_005736_070 |
0.3409355465 |
transcription factor |
TF Family: GRAS |
PREDICTED: scarecrow-like protein 1 [Jatropha curcas] |
| 11 |
Hb_124207_010 |
0.3423270006 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 12 |
Hb_000523_060 |
0.3457567316 |
- |
- |
- |
| 13 |
Hb_000510_350 |
0.3457765577 |
- |
- |
Putative polyprotein [Oryza sativa Japonica Group] |
| 14 |
Hb_012553_010 |
0.3460491432 |
- |
- |
hypothetical protein JCGZ_10552 [Jatropha curcas] |
| 15 |
Hb_002787_100 |
0.3461591811 |
- |
- |
PREDICTED: uncharacterized protein LOC105637209 [Jatropha curcas] |
| 16 |
Hb_000291_020 |
0.3462672579 |
- |
- |
hypothetical protein POPTR_0019s06080g [Populus trichocarpa] |
| 17 |
Hb_002035_070 |
0.3549853178 |
- |
- |
hypothetical protein CICLE_v10033569mg [Citrus clementina] |
| 18 |
Hb_000878_150 |
0.3553513638 |
- |
- |
Gibberellin receptor GID1, putative [Ricinus communis] |
| 19 |
Hb_004460_040 |
0.3563578075 |
- |
- |
- |
| 20 |
Hb_108802_010 |
0.357871451 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |