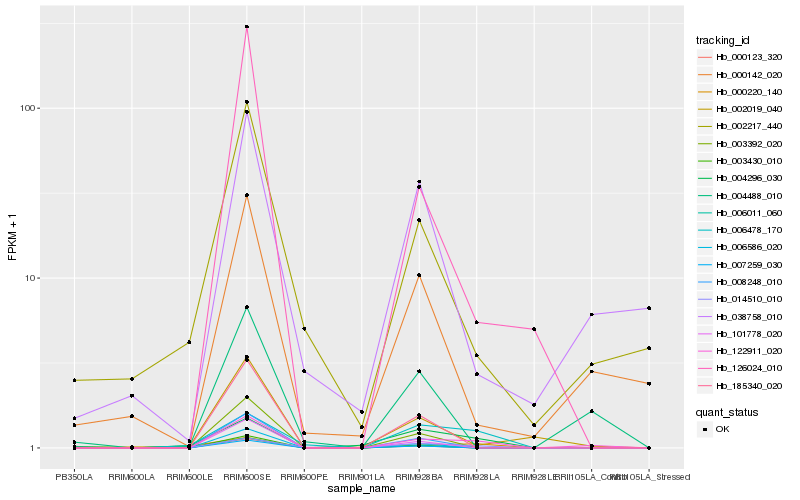
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_122911_020 |
0.0 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 2 |
Hb_126024_010 |
0.2802348402 |
- |
- |
PREDICTED: probable aldo-keto reductase 1 [Populus euphratica] |
| 3 |
Hb_000142_020 |
0.2809016513 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 4 |
Hb_004488_010 |
0.2826872524 |
- |
- |
hypothetical protein CISIN_1g019168mg [Citrus sinensis] |
| 5 |
Hb_014510_010 |
0.284033411 |
- |
- |
hypothetical protein JCGZ_08917 [Jatropha curcas] |
| 6 |
Hb_038758_010 |
0.2862007229 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 7 |
Hb_002019_040 |
0.2884413241 |
- |
- |
hypothetical protein PRUPE_ppb016160mg, partial [Prunus persica] |
| 8 |
Hb_006478_170 |
0.2884914009 |
- |
- |
hypothetical protein POPTR_0005s23360g [Populus trichocarpa] |
| 9 |
Hb_000220_140 |
0.289504822 |
- |
- |
palmitoyl-acyl carrier protein thioesterase [Ricinus communis] |
| 10 |
Hb_002217_440 |
0.2931791558 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 11 |
Hb_008248_010 |
0.2967087903 |
- |
- |
- |
| 12 |
Hb_185340_020 |
0.2967655316 |
- |
- |
- |
| 13 |
Hb_007259_030 |
0.2967710986 |
- |
- |
- |
| 14 |
Hb_006011_060 |
0.2968403071 |
- |
- |
- |
| 15 |
Hb_003392_020 |
0.296860715 |
- |
- |
PREDICTED: uncharacterized protein LOC105650609 [Jatropha curcas] |
| 16 |
Hb_003430_010 |
0.297129887 |
- |
- |
xyloglucan endotransglucosylase/hydrolase protein 33 precursor [Populus trichocarpa] |
| 17 |
Hb_000123_320 |
0.297209985 |
- |
- |
PREDICTED: MADS-box protein FLOWERING LOCUS C-like [Jatropha curcas] |
| 18 |
Hb_006586_020 |
0.2977010248 |
- |
- |
hypothetical protein [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_004296_030 |
0.297955833 |
- |
- |
PREDICTED: gamma-tubulin complex component 3 [Jatropha curcas] |
| 20 |
Hb_101778_020 |
0.298063956 |
transcription factor |
TF Family: SRS |
conserved hypothetical protein [Ricinus communis] |