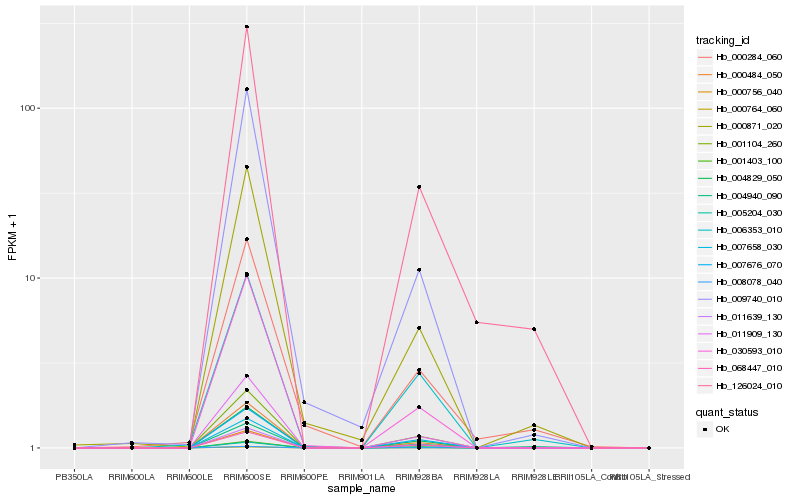
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_126024_010 |
0.0 |
- |
- |
PREDICTED: probable aldo-keto reductase 1 [Populus euphratica] |
| 2 |
Hb_011909_130 |
0.1122103553 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB86 [Populus euphratica] |
| 3 |
Hb_011639_130 |
0.1143719741 |
- |
- |
mads box protein, putative [Ricinus communis] |
| 4 |
Hb_004829_050 |
0.1146818112 |
- |
- |
hypothetical protein TRIUR3_09654 [Triticum urartu] |
| 5 |
Hb_000484_050 |
0.1154222346 |
- |
- |
hypothetical protein CICLE_v10028549mg [Citrus clementina] |
| 6 |
Hb_001403_100 |
0.116694398 |
- |
- |
hypothetical protein CICLE_v10017457mg, partial [Citrus clementina] |
| 7 |
Hb_001104_260 |
0.1168249921 |
- |
- |
hypothetical protein JCGZ_23628 [Jatropha curcas] |
| 8 |
Hb_007658_030 |
0.1174878429 |
- |
- |
PREDICTED: lupeol synthase-like isoform X1 [Populus euphratica] |
| 9 |
Hb_000871_020 |
0.1175308395 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 10 |
Hb_000284_060 |
0.1183378153 |
desease resistance |
Gene Name: NB-ARC |
disease resistance protein (TIR-NBS-LRR class), putative [Medicago truncatula] |
| 11 |
Hb_006353_010 |
0.118524044 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105634775 [Jatropha curcas] |
| 12 |
Hb_005204_030 |
0.1201499072 |
- |
- |
PREDICTED: L-type lectin-domain containing receptor kinase IX.1-like [Jatropha curcas] |
| 13 |
Hb_008078_040 |
0.1217475741 |
- |
- |
PREDICTED: uncharacterized protein LOC105764710 isoform X3 [Gossypium raimondii] |
| 14 |
Hb_000756_040 |
0.1222945348 |
- |
- |
anthocyanidin 3-O-glucosyltransferase [Hevea brasiliensis] |
| 15 |
Hb_000764_060 |
0.1239984274 |
- |
- |
PREDICTED: uncharacterized protein LOC101491058 [Cicer arietinum] |
| 16 |
Hb_007676_070 |
0.1246373897 |
- |
- |
PREDICTED: omega-6 fatty acid desaturase, endoplasmic reticulum isozyme 1-like [Populus euphratica] |
| 17 |
Hb_004940_090 |
0.1250906715 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 18 |
Hb_030593_010 |
0.1252944185 |
- |
- |
hypothetical protein JCGZ_06063 [Jatropha curcas] |
| 19 |
Hb_068447_010 |
0.1263093267 |
- |
- |
protein with unknown function [Ricinus communis] |
| 20 |
Hb_009740_010 |
0.1267289107 |
- |
- |
receptor-kinase, putative [Ricinus communis] |