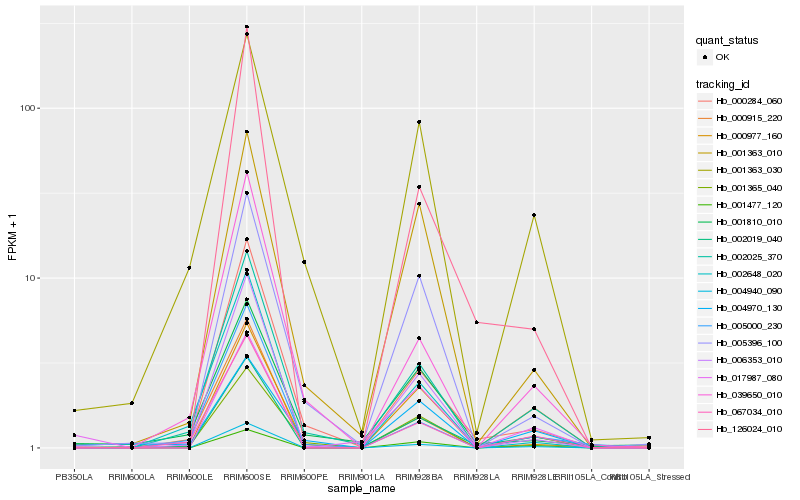
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002019_040 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppb016160mg, partial [Prunus persica] |
| 2 |
Hb_002025_370 |
0.1528692996 |
- |
- |
monooxygenase, putative [Ricinus communis] |
| 3 |
Hb_001477_120 |
0.1549753271 |
- |
- |
hypothetical protein JCGZ_25721 [Jatropha curcas] |
| 4 |
Hb_004940_090 |
0.1625751223 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 5 |
Hb_067034_010 |
0.1631713228 |
- |
- |
PREDICTED: vacuolar cation/proton exchanger 3-like [Jatropha curcas] |
| 6 |
Hb_006353_010 |
0.16353406 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105634775 [Jatropha curcas] |
| 7 |
Hb_000284_060 |
0.1696634192 |
desease resistance |
Gene Name: NB-ARC |
disease resistance protein (TIR-NBS-LRR class), putative [Medicago truncatula] |
| 8 |
Hb_126024_010 |
0.1700769219 |
- |
- |
PREDICTED: probable aldo-keto reductase 1 [Populus euphratica] |
| 9 |
Hb_001365_040 |
0.1754732357 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 isoform X2 [Populus euphratica] |
| 10 |
Hb_001363_010 |
0.1784917591 |
- |
- |
hypothetical protein CISIN_1g027763mg [Citrus sinensis] |
| 11 |
Hb_001810_010 |
0.1796000556 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Nelumbo nucifera] |
| 12 |
Hb_001363_030 |
0.1797443903 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F2-like [Populus euphratica] |
| 13 |
Hb_000977_160 |
0.1800627208 |
- |
- |
Serine-threonine protein kinase, plant-type, putative [Theobroma cacao] |
| 14 |
Hb_005396_100 |
0.1875786635 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_017987_080 |
0.1876997904 |
- |
- |
Uncharacterized protein TCM_001832 [Theobroma cacao] |
| 16 |
Hb_000915_220 |
0.1907248442 |
- |
- |
Lactoylglutathione lyase / glyoxalase I family protein [Theobroma cacao] |
| 17 |
Hb_002648_020 |
0.1954278729 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 330-like [Populus euphratica] |
| 18 |
Hb_005000_230 |
0.1963493191 |
transcription factor |
TF Family: bHLH |
PREDICTED: putative transcription factor bHLH041 [Jatropha curcas] |
| 19 |
Hb_004970_130 |
0.1973431252 |
- |
- |
PREDICTED: uncharacterized protein LOC105633891 [Jatropha curcas] |
| 20 |
Hb_039650_010 |
0.1988428728 |
- |
- |
cellulose synthase-like protein [Populus tomentosa] |