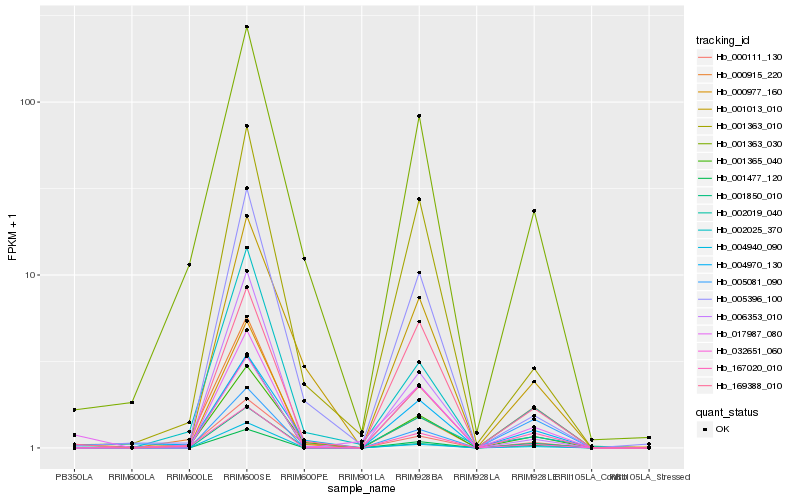
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001477_120 |
0.0 |
- |
- |
hypothetical protein JCGZ_25721 [Jatropha curcas] |
| 2 |
Hb_001365_040 |
0.1421030203 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 isoform X2 [Populus euphratica] |
| 3 |
Hb_169388_010 |
0.151489508 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 4 |
Hb_002019_040 |
0.1549753271 |
- |
- |
hypothetical protein PRUPE_ppb016160mg, partial [Prunus persica] |
| 5 |
Hb_167020_010 |
0.1619595718 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 6 |
Hb_017987_080 |
0.1642295022 |
- |
- |
Uncharacterized protein TCM_001832 [Theobroma cacao] |
| 7 |
Hb_001363_010 |
0.1652990386 |
- |
- |
hypothetical protein CISIN_1g027763mg [Citrus sinensis] |
| 8 |
Hb_004940_090 |
0.1688048589 |
- |
- |
PREDICTED: putative receptor-like protein kinase At3g47110 [Jatropha curcas] |
| 9 |
Hb_002025_370 |
0.1761773801 |
- |
- |
monooxygenase, putative [Ricinus communis] |
| 10 |
Hb_001363_030 |
0.1792845646 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F2-like [Populus euphratica] |
| 11 |
Hb_000977_160 |
0.1833202176 |
- |
- |
Serine-threonine protein kinase, plant-type, putative [Theobroma cacao] |
| 12 |
Hb_004970_130 |
0.183413225 |
- |
- |
PREDICTED: uncharacterized protein LOC105633891 [Jatropha curcas] |
| 13 |
Hb_006353_010 |
0.183777508 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105634775 [Jatropha curcas] |
| 14 |
Hb_005396_100 |
0.1849902439 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005081_090 |
0.1861725131 |
- |
- |
- |
| 16 |
Hb_000915_220 |
0.1882618892 |
- |
- |
Lactoylglutathione lyase / glyoxalase I family protein [Theobroma cacao] |
| 17 |
Hb_001013_010 |
0.1930686437 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 18 |
Hb_032651_060 |
0.1934426373 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 19 |
Hb_000111_130 |
0.2001353304 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 20 |
Hb_001850_010 |
0.2005193844 |
- |
- |
PREDICTED: cytochrome P450 71B34-like [Jatropha curcas] |