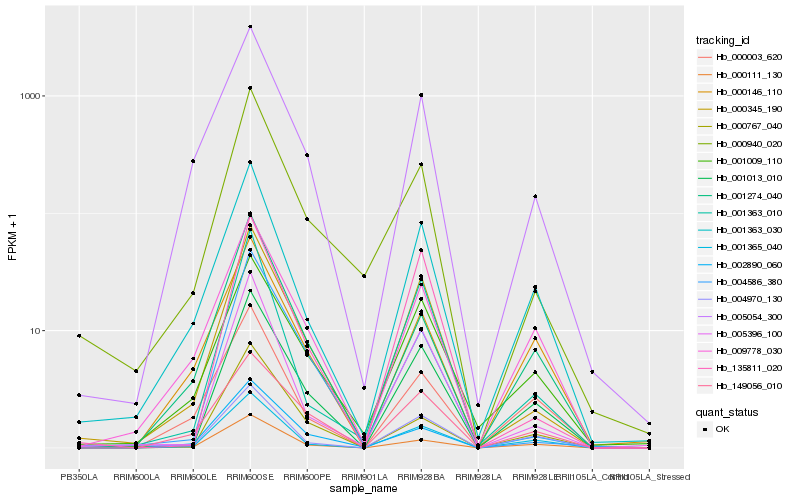
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001013_010 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_001365_040 |
0.0990358749 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 isoform X2 [Populus euphratica] |
| 3 |
Hb_000111_130 |
0.1166450129 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 4 |
Hb_000345_190 |
0.1204398528 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0005s02730g [Populus trichocarpa] |
| 5 |
Hb_001363_030 |
0.1284873399 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F2-like [Populus euphratica] |
| 6 |
Hb_005396_100 |
0.1330740544 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_009778_030 |
0.1333742171 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP23-like [Populus euphratica] |
| 8 |
Hb_001009_110 |
0.1360610965 |
- |
- |
PREDICTED: uncharacterized protein LOC105640232 [Jatropha curcas] |
| 9 |
Hb_004970_130 |
0.1403385819 |
- |
- |
PREDICTED: uncharacterized protein LOC105633891 [Jatropha curcas] |
| 10 |
Hb_149056_010 |
0.1417035583 |
- |
- |
hypothetical protein CICLE_v10016664mg [Citrus clementina] |
| 11 |
Hb_002890_060 |
0.1420206719 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA1 [Eucalyptus grandis] |
| 12 |
Hb_001363_010 |
0.1461721821 |
- |
- |
hypothetical protein CISIN_1g027763mg [Citrus sinensis] |
| 13 |
Hb_000767_040 |
0.1463074282 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 14 |
Hb_135811_020 |
0.1548585678 |
- |
- |
hypothetical protein JCGZ_19965 [Jatropha curcas] |
| 15 |
Hb_000940_020 |
0.1555718152 |
- |
- |
hypothetical protein POPTR_0004s17500g [Populus trichocarpa] |
| 16 |
Hb_000003_620 |
0.157432459 |
- |
- |
PREDICTED: ABC transporter G family member 29-like [Jatropha curcas] |
| 17 |
Hb_001274_040 |
0.1576622923 |
- |
- |
PREDICTED: uncharacterized protein LOC105636321 [Jatropha curcas] |
| 18 |
Hb_005054_300 |
0.1582455143 |
- |
- |
aquaporin [Hevea brasiliensis] |
| 19 |
Hb_000146_110 |
0.1599599708 |
- |
- |
resveratrol/hydroxycinnamic acid O-glucosyltransferase [Vitis labrusca] |
| 20 |
Hb_004586_380 |
0.161928256 |
- |
- |
purine transporter, putative [Ricinus communis] |