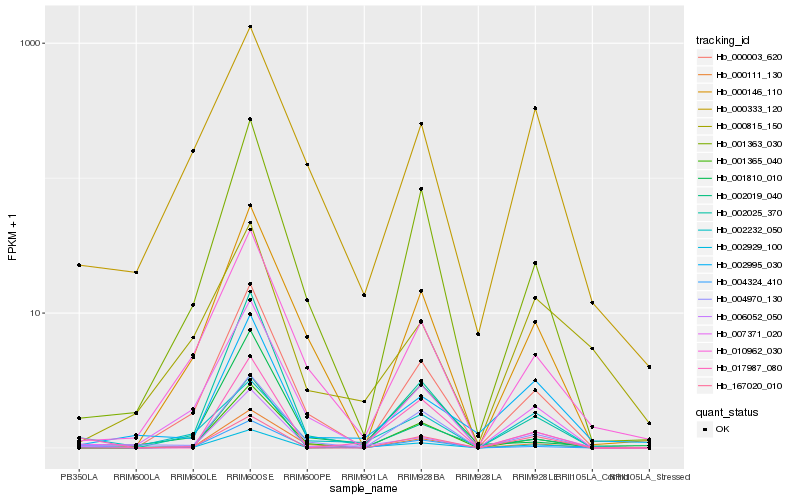
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002929_100 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 2 |
Hb_006052_050 |
0.1622911001 |
- |
- |
hypothetical protein EUGRSUZ_A00568 [Eucalyptus grandis] |
| 3 |
Hb_017987_080 |
0.1912750356 |
- |
- |
Uncharacterized protein TCM_001832 [Theobroma cacao] |
| 4 |
Hb_004324_410 |
0.2045849839 |
- |
- |
hypothetical protein JCGZ_26025 [Jatropha curcas] |
| 5 |
Hb_002019_040 |
0.2145811238 |
- |
- |
hypothetical protein PRUPE_ppb016160mg, partial [Prunus persica] |
| 6 |
Hb_002232_050 |
0.2191523381 |
- |
- |
PREDICTED: uncharacterized protein LOC104214925 [Nicotiana sylvestris] |
| 7 |
Hb_001363_030 |
0.2201084646 |
- |
- |
PREDICTED: UDP-glycosyltransferase 74F2-like [Populus euphratica] |
| 8 |
Hb_002995_030 |
0.2201954059 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_004970_130 |
0.2205447663 |
- |
- |
PREDICTED: uncharacterized protein LOC105633891 [Jatropha curcas] |
| 10 |
Hb_010962_030 |
0.225387582 |
- |
- |
kinase, putative [Ricinus communis] |
| 11 |
Hb_000003_620 |
0.227113207 |
- |
- |
PREDICTED: ABC transporter G family member 29-like [Jatropha curcas] |
| 12 |
Hb_000815_150 |
0.2290887507 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 13 |
Hb_001365_040 |
0.2306340921 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At1g05700 isoform X2 [Populus euphratica] |
| 14 |
Hb_002025_370 |
0.231061597 |
- |
- |
monooxygenase, putative [Ricinus communis] |
| 15 |
Hb_000333_120 |
0.2345466755 |
- |
- |
PREDICTED: thiol protease aleurain-like [Jatropha curcas] |
| 16 |
Hb_007371_020 |
0.2378319614 |
- |
- |
PREDICTED: ent-kaurenoic acid oxidase 1-like isoform X1 [Jatropha curcas] |
| 17 |
Hb_001810_010 |
0.2424154815 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 1 [Nelumbo nucifera] |
| 18 |
Hb_167020_010 |
0.2436974944 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_000111_130 |
0.2438625442 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 20 |
Hb_000146_110 |
0.2477931677 |
- |
- |
resveratrol/hydroxycinnamic acid O-glucosyltransferase [Vitis labrusca] |